Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size
- PMID: 28472462
- PMCID: PMC5853923
- DOI: 10.1093/jxb/erx147
Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size
Abstract
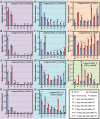
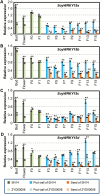
Soybean (Glycine max) probably originated from the wild soybean (Glycine soja). Glycine max has a significantly larger seed size, but the underlying genomic changes are largely unknown. Candidate regulatory genes were preliminarily proposed by data co-localizing RNA sequencing with the quantitative loci (QTLs) for seed size. The soybean gene locus SoyWRKY15a and its orthologous genes from G. max (GmWRKY15a) and G. soja (GsWRKY15a) were analyzed in detail. The coding sequences were nearly identical between the two orthologs, but GmWRKY15a was significantly more highly expressed than GsWRKY15a. Four haplotypes (H1-H4) were found and they varied in the size of a CT-core microsatellite locus in the 5'-untranslated region of this gene. H1 (with six CT-repeats) was the only allelic version found in G. max, while H3 (with five CT-repeats) was the dominant G. soja allele. Differential expression of this gene in soybean pods was correlated with CT-repeat variation, and manipulation of the CT copy number altered the reporter gene expression, suggesting a regulatory role for the simple sequence repeats. Seed weight of wild soybeans harboring H1 was significantly greater than that of soybeans having haplotypes H2, H3, or H4, and seed weight was correlated with gene expression, suggesting the influence of GsWRKY15a in controlling seed size. However, the seed size might be refractory to increased SoyWRKY15a expression in cultivated soybeans. The evolutionary significance of SoyWRKY15a variation in soybean seed domestication is discussed.
Keywords: CT-rich regulatory motif; WRKY gene; domestication; expression variation; seed size; soybean.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures




References
-
- Alexandrova KS, Conger BV. 2002. Isolation of two somatic embryogenesis-related genes from orchardgrass (Dactylis glomerata). Plant Science 162, 301–307.
-
- Audic S, Claverie JM. 1997. The significance of digital gene expression profiles. Genome Research 7, 986–995. - PubMed
-
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 29, 1165–1188.
-
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. - PubMed
-
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

