Predicting genome terminus sequences of Bacillus cereus-group bacteriophage using next generation sequencing data
- PMID: 28472946
- PMCID: PMC5418689
- DOI: 10.1186/s12864-017-3744-0
Predicting genome terminus sequences of Bacillus cereus-group bacteriophage using next generation sequencing data
Abstract
Background: Most tailed bacteriophages (phages) feature linear dsDNA genomes. Characterizing novel phages requires an understanding of complete genome sequences, including the definition of genome physical ends.
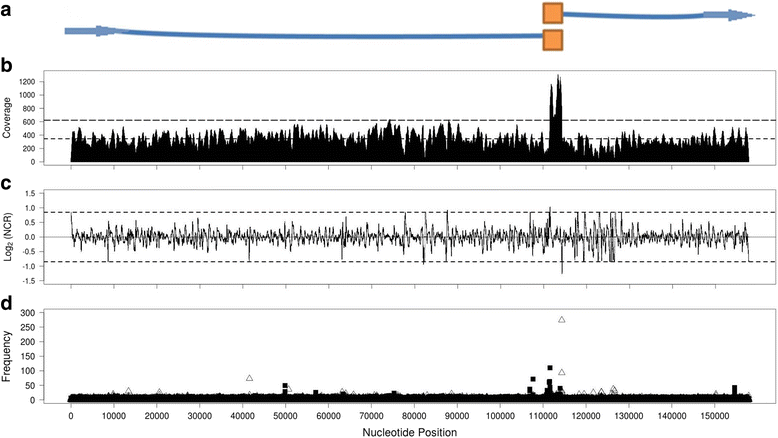
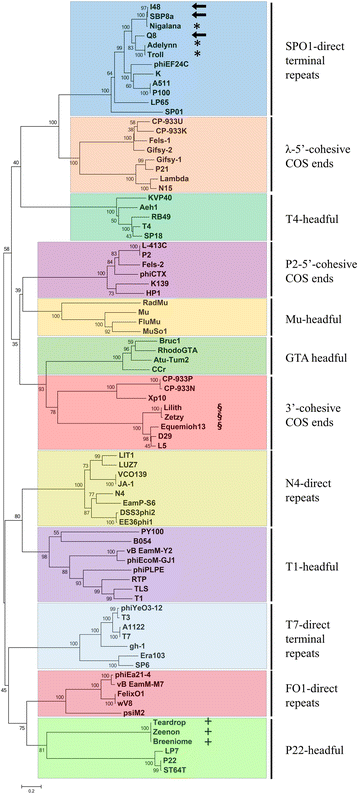
Result: We sequenced 48 Bacillus cereus phage isolates and analyzed Next-generation sequencing (NGS) data to resolve the genome configuration of these novel phages. Most assembled contigs featured reads that mapped to both contig ends and formed circularized contigs. Independent assemblies of 31 nearly identical I48-like Bacillus phage isolates allowed us to observe that the assembly programs tended to produce random cleavage on circularized contigs. However, currently available assemblers were not capable of reporting the underlying phage genome configuration from sequence data. To identify the genome configuration of sequenced phage in silico, a terminus prediction method was developed by means of 'neighboring coverage ratios' and 'read edge frequencies' from read alignment files. Termini were confirmed by primer walking and supported by phylogenetic inference of large DNA terminase protein sequences.
Conclusions: The Terminus package using phage NGS data along with the contig circularity could efficiently identify the proximal positions of phage genome terminus. Complete phage genome sequences allow a proposed characterization of the potential packaging mechanisms and more precise genome annotation.
Keywords: Bacteriophage; Direct terminal repeat; Genome packaging mechanisms; Neighboring coverage ratio; Phage genome configuration; Read edge frequency; Terminus prediction.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

