Tracking microbial colonization in fecal microbiota transplantation experiments via genome-resolved metagenomics
- PMID: 28473000
- PMCID: PMC5418705
- DOI: 10.1186/s40168-017-0270-x
Tracking microbial colonization in fecal microbiota transplantation experiments via genome-resolved metagenomics
Abstract
Background: Fecal microbiota transplantation (FMT) is an effective treatment for recurrent Clostridium difficile infection and shows promise for treating other medical conditions associated with intestinal dysbioses. However, we lack a sufficient understanding of which microbial populations successfully colonize the recipient gut, and the widely used approaches to study the microbial ecology of FMT experiments fail to provide enough resolution to identify populations that are likely responsible for FMT-derived benefits.
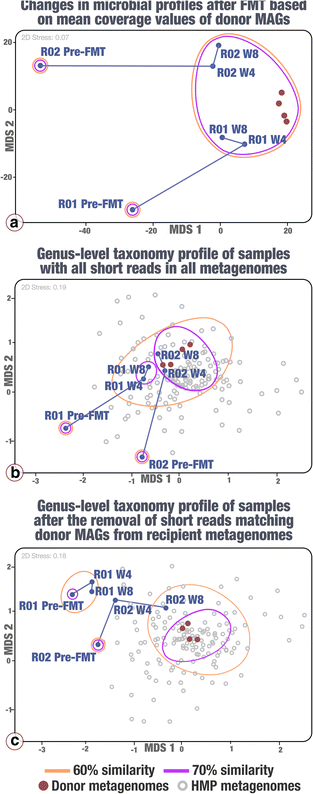
Methods: We used shotgun metagenomics together with assembly and binning strategies to reconstruct metagenome-assembled genomes (MAGs) from fecal samples of a single FMT donor. We then used metagenomic mapping to track the occurrence and distribution patterns of donor MAGs in two FMT recipients.
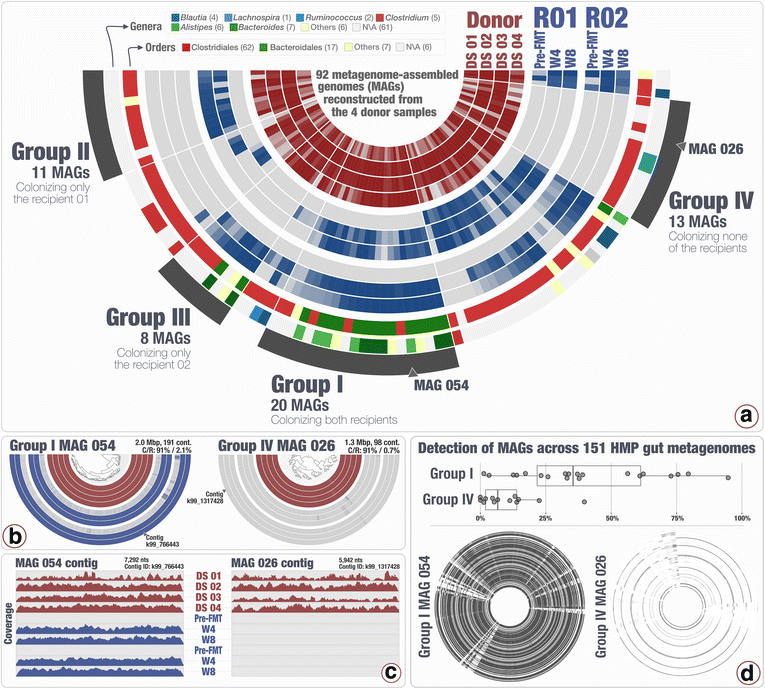
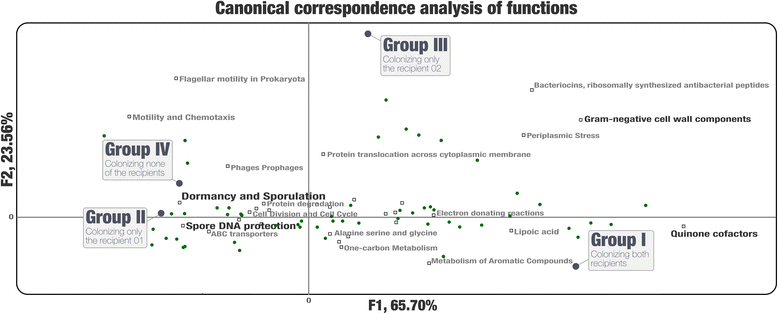
Results: Our analyses revealed that 22% of the 92 highly complete bacterial MAGs that we identified from the donor successfully colonized and remained abundant in two recipients for at least 8 weeks. Most MAGs with a high colonization rate belonged to the order Bacteroidales. The vast majority of those that lacked evidence of colonization belonged to the order Clostridiales, and colonization success was negatively correlated with the number of genes related to sporulation. Our analysis of 151 publicly available gut metagenomes showed that the donor MAGs that colonized both recipients were prevalent, and the ones that colonized neither were rare across the participants of the Human Microbiome Project. Although our dataset showed a link between taxonomy and the colonization ability of a given MAG, we also identified MAGs that belong to the same taxon with different colonization properties, highlighting the importance of an appropriate level of resolution to explore the functional basis of colonization and to identify targets for cultivation, hypothesis generation, and testing in model systems.
Conclusions: The analytical strategy adopted in our study can provide genomic insights into bacterial populations that may be critical to the efficacy of FMT due to their success in gut colonization and metabolic properties, and guide cultivation efforts to investigate mechanistic underpinnings of this procedure beyond associations.
Keywords: Colonization; Fecal microbiota transplantation; Metagenome-assembled genomes; Metagenomics.
Figures
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

