Mapping the human DC lineage through the integration of high-dimensional techniques
- PMID: 28473638
- PMCID: PMC7611082
- DOI: 10.1126/science.aag3009
Mapping the human DC lineage through the integration of high-dimensional techniques
Abstract
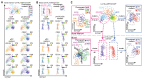
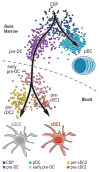
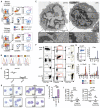
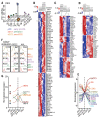
Dendritic cells (DC) are professional antigen-presenting cells that orchestrate immune responses. The human DC population comprises two main functionally specialized lineages, whose origins and differentiation pathways remain incompletely defined. Here, we combine two high-dimensional technologies-single-cell messenger RNA sequencing (scmRNAseq) and cytometry by time-of-flight (CyTOF)-to identify human blood CD123+CD33+CD45RA+ DC precursors (pre-DC). Pre-DC share surface markers with plasmacytoid DC (pDC) but have distinct functional properties that were previously attributed to pDC. Tracing the differentiation of DC from the bone marrow to the peripheral blood revealed that the pre-DC compartment contains distinct lineage-committed subpopulations, including one early uncommitted CD123high pre-DC subset and two CD45RA+CD123low lineage-committed subsets exhibiting functional differences. The discovery of multiple committed pre-DC populations opens promising new avenues for the therapeutic exploitation of DC subset-specific targeting.
Copyright © 2017, American Association for the Advancement of Science.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

