Mechanical Strain Promotes Oligodendrocyte Differentiation by Global Changes of Gene Expression
- PMID: 28473753
- PMCID: PMC5397481
- DOI: 10.3389/fncel.2017.00093
Mechanical Strain Promotes Oligodendrocyte Differentiation by Global Changes of Gene Expression
Abstract
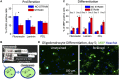
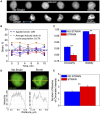
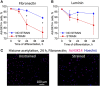
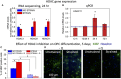
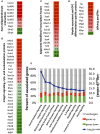
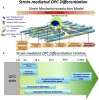
Differentiation of oligodendrocyte progenitor cells (OPC) to oligodendrocytes and subsequent axon myelination are critical steps in vertebrate central nervous system (CNS) development and regeneration. Growing evidence supports the significance of mechanical factors in oligodendrocyte biology. Here, we explore the effect of mechanical strains within physiological range on OPC proliferation and differentiation, and strain-associated changes in chromatin structure, epigenetics, and gene expression. Sustained tensile strain of 10-15% inhibited OPC proliferation and promoted differentiation into oligodendrocytes. This response to strain required specific interactions of OPCs with extracellular matrix ligands. Applied strain induced changes in nuclear shape, chromatin organization, and resulted in enhanced histone deacetylation, consistent with increased oligodendrocyte differentiation. This response was concurrent with increased mRNA levels of the epigenetic modifier histone deacetylase Hdac11. Inhibition of HDAC proteins eliminated the strain-mediated increase of OPC differentiation, demonstrating a role of HDACs in mechanotransduction of strain to chromatin. RNA sequencing revealed global changes in gene expression associated with strain. Specifically, expression of multiple genes associated with oligodendrocyte differentiation and axon-oligodendrocyte interactions was increased, including cell surface ligands (Ncam, ephrins), cyto- and nucleo-skeleton genes (Fyn, actinins, myosin, nesprin, Sun1), transcription factors (Sox10, Zfp191, Nkx2.2), and myelin genes (Cnp, Plp, Mag). These findings show how mechanical strain can be transmitted to the nucleus to promote oligodendrocyte differentiation, and identify the global landscape of signaling pathways involved in mechanotransduction. These data provide a source of potential new therapeutic avenues to enhance OPC differentiation in vivo.
Keywords: cell nucleus shape; chromatin remodeling; mechanical strain; mechanotransduction; multiple sclerosis (MS); oligodendrocyte differentiation; oligodendrocyte precursor cell (OPC); oligodendrocytes.
Figures
References
-
- Baer A. S., Syed Y. A., Kang S. U., Mitteregger D., Vig R., ffrench-Constant C., et al. (2009). Myelin-mediated inhibition of oligodendrocyte precursor differentiation can be overcome by pharmacological modulation of Fyn-RhoA and protein kinase C signalling. Brain 132(Pt 2), 465–481. 10.1093/brain/awn334 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

