Reactivation and Lytic Replication of Kaposi's Sarcoma-Associated Herpesvirus: An Update
- PMID: 28473805
- PMCID: PMC5397509
- DOI: 10.3389/fmicb.2017.00613
Reactivation and Lytic Replication of Kaposi's Sarcoma-Associated Herpesvirus: An Update
Abstract
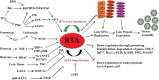
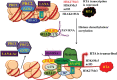
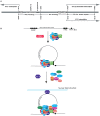
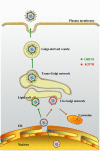
The life cycle of Kaposi's sarcoma-associated herpesvirus (KSHV) consists of two phases, latent and lytic. The virus establishes latency as a strategy for avoiding host immune surveillance and fusing symbiotically with the host for lifetime persistent infection. However, latency can be disrupted and KSHV is reactivated for entry into the lytic replication. Viral lytic replication is crucial for efficient dissemination from its long-term reservoir to the sites of disease and for the spread of the virus to new hosts. The balance of these two phases in the KSHV life cycle is important for both the virus and the host and control of the switch between these two phases is extremely complex. Various environmental factors such as oxidative stress, hypoxia, and certain chemicals have been shown to switch KSHV from latency to lytic reactivation. Immunosuppression, unbalanced inflammatory cytokines, and other viral co-infections also lead to the reactivation of KSHV. This review article summarizes the current understanding of the initiation and regulation of KSHV reactivation and the mechanisms underlying the process of viral lytic replication. In particular, the central role of an immediate-early gene product RTA in KSHV reactivation has been extensively investigated. These studies revealed multiple layers of regulation in activation of RTA as well as the multifunctional roles of RTA in the lytic replication cascade. Epigenetic regulation is known as a critical layer of control for the switch of KSHV between latency and lytic replication. The viral non-coding RNA, PAN, was demonstrated to play a central role in the epigenetic regulation by serving as a guide RNA that brought chromatin remodeling enzymes to the promoters of RTA and other lytic genes. In addition, a novel dimension of regulation by microPeptides emerged and has been shown to regulate RTA expression at the protein level. Overall, extensive investigation of KSHV reactivation and lytic replication has revealed a sophisticated regulation network that controls the important events in KSHV life cycle.
Keywords: Kaposi’s sarcoma-associated herpesvirus (KSHV); Rta; human herpesvirus 8 (HHV-8); lytic replication; viral reactivation.
Figures

Similar articles
-
Genome-Wide Identification of Direct RTA Targets Reveals Key Host Factors for Kaposi's Sarcoma-Associated Herpesvirus Lytic Reactivation.J Virol. 2019 Feb 19;93(5):e01978-18. doi: 10.1128/JVI.01978-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541837 Free PMC article.
-
Activation of Kaposi's sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: identification of sirtuin 1 as a regulator of the KSHV life cycle.J Virol. 2014 Jun;88(11):6355-67. doi: 10.1128/JVI.00219-14. Epub 2014 Mar 26. J Virol. 2014. PMID: 24672028 Free PMC article.
-
ZIC2 Is Essential for Maintenance of Latency and Is a Target of an Immediate Early Protein during Kaposi's Sarcoma-Associated Herpesvirus Lytic Reactivation.J Virol. 2017 Oct 13;91(21):e00980-17. doi: 10.1128/JVI.00980-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28835494 Free PMC article.
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
-
Molecular biology of KSHV lytic reactivation.Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review.
Cited by
-
Viruses and Skin Cancer.Int J Mol Sci. 2021 May 20;22(10):5399. doi: 10.3390/ijms22105399. Int J Mol Sci. 2021. PMID: 34065594 Free PMC article. Review.
-
KSHV-encoded LANA bypasses transcriptional block through the stabilization of RNA Pol II in hypoxia.mBio. 2024 Jan 16;15(1):e0277423. doi: 10.1128/mbio.02774-23. Epub 2023 Dec 14. mBio. 2024. PMID: 38095447 Free PMC article.
-
PAX5 functions as a tumor suppressor by RB-E2F-mediated cell cycle arrest in Kaposi sarcoma-associated herpesvirus-infected primary effusion lymphoma.Neoplasia. 2024 Oct;56:101035. doi: 10.1016/j.neo.2024.101035. Epub 2024 Aug 2. Neoplasia. 2024. PMID: 39096792 Free PMC article.
-
Molecular Mechanisms of Flavonoids against Tumor Gamma-Herpesviruses and Their Correlated Cancers-A Focus on EBV and KSHV Life Cycles and Carcinogenesis.Int J Mol Sci. 2022 Dec 23;24(1):247. doi: 10.3390/ijms24010247. Int J Mol Sci. 2022. PMID: 36613688 Free PMC article. Review.
-
Oral Shedding of an Oncogenic Virus Alters the Oral Microbiome in HIV+ Patients.Front Microbiol. 2022 Apr 19;13:882520. doi: 10.3389/fmicb.2022.882520. eCollection 2022. Front Microbiol. 2022. PMID: 35516440 Free PMC article.
References
-
- Anders D. G., Kerry J. A., Pari G. S. (2007). “DNA synthesis and late viral gene expression,” in Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, eds Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., et al. (Cambridge: Cambridge University Press; ), 19. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

