Discrimination of Self and Non-Self Ribonucleic Acids
- PMID: 28475460
- PMCID: PMC5439445
- DOI: 10.1089/jir.2016.0092
Discrimination of Self and Non-Self Ribonucleic Acids
Abstract
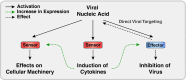
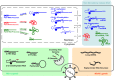
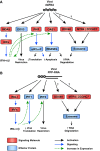
Most virus infections are controlled through the innate and adaptive immune system. A surprisingly limited number of so-called pattern recognition receptors (PRRs) have the ability to sense a large variety of virus infections. The reason for the broad activity of PRRs lies in the ability to recognize viral nucleic acids. These nucleic acids lack signatures that are present in cytoplasmic cellular nucleic acids and thereby marking them as pathogen-derived. Accumulating evidence suggests that these signatures, which are predominantly sensed by a class of PRRs called retinoic acid-inducible gene I (RIG-I)-like receptors and other proteins, are not unique to viruses but rather resemble immature forms of cellular ribonucleic acids generated by cellular polymerases. RIG-I-like receptors, and other cellular antiviral proteins, may therefore have mainly evolved to sense nonprocessed nucleic acids typically generated by primitive organisms and pathogens. This capability has not only implications on induction of antiviral immunity but also on the function of cellular proteins to handle self-derived RNA with stimulatory potential.
Keywords: MDA5; PRR; RIG-I; antiviral mechanisms; interferon; ribonucleic acid sensing.
Conflict of interest statement
No competing financial interests exist.
Figures
References
-
- Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA. 2012. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J 31(21):4153–4164 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

