An Immune Atlas of Clear Cell Renal Cell Carcinoma
- PMID: 28475899
- PMCID: PMC5422211
- DOI: 10.1016/j.cell.2017.04.016
An Immune Atlas of Clear Cell Renal Cell Carcinoma
Abstract
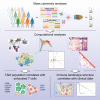
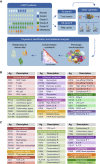
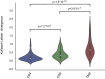
Immune cells in the tumor microenvironment modulate cancer progression and are attractive therapeutic targets. Macrophages and T cells are key components of the microenvironment, yet their phenotypes and relationships in this ecosystem and to clinical outcomes are ill defined. We used mass cytometry with extensive antibody panels to perform in-depth immune profiling of samples from 73 clear cell renal cell carcinoma (ccRCC) patients and five healthy controls. In 3.5 million measured cells, we identified 17 tumor-associated macrophage phenotypes, 22 T cell phenotypes, and a distinct immune composition correlated with progression-free survival, thereby presenting an in-depth human atlas of the immune tumor microenvironment in this disease. This study revealed potential biomarkers and targets for immunotherapy development and validated tools that can be used for immune profiling of other tumor types.
Keywords: T cell; clear cell renal cell carcinoma; immune cell atlas; immunosuppression; macrophage; mass cytometry; single-cell analysis.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures












Comment in
-
Heavy Metal Enlightens Tumor Immunity.Cell. 2017 May 4;169(4):567-569. doi: 10.1016/j.cell.2017.04.017. Cell. 2017. PMID: 28475889 Free PMC article.
-
Cell maps reveal fresh details on how the immune system fights cancer.Nature. 2017 May 4;545(7653):143. doi: 10.1038/nature.2017.21931. Nature. 2017. PMID: 28492266 No abstract available.
-
Tumour immunology: Immune atlas sheds light on anticancer responses.Nat Rev Immunol. 2017 May 26;17(6):347. doi: 10.1038/nri.2017.59. Nat Rev Immunol. 2017. PMID: 28548134 No abstract available.
-
Heavy Metal to Rock the Immune Infiltrate.Trends Immunol. 2017 Aug;38(8):539-541. doi: 10.1016/j.it.2017.05.007. Epub 2017 Jun 9. Trends Immunol. 2017. PMID: 28602618
-
Unmassked: Single-Cell Profiling of Immune Cell Populations in Tumors.Mol Ther. 2017 Aug 2;25(8):1745-1747. doi: 10.1016/j.ymthe.2017.06.024. Epub 2017 Jul 4. Mol Ther. 2017. PMID: 28684257 Free PMC article. No abstract available.
-
Defining Renal Neoplastic Disease, One Cell at a Time: Mass Cytometry, a New Tool for the Study of Kidney Biology and Disease.Am J Kidney Dis. 2017 Dec;70(6):758-761. doi: 10.1053/j.ajkd.2017.08.009. Epub 2017 Oct 12. Am J Kidney Dis. 2017. PMID: 29031857 No abstract available.
References
-
- Angerer P., Haghverdi L., Büttner M., Theis F.J., Marr C., Buettner F. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. - PubMed
-
- Asano K., Nabeyama A., Miyake Y., Qiu C.H., Kurita A., Tomura M., Kanagawa O., Fujii S., Tanaka M. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity. 2011;34:85–95. - PubMed
-
- Bandura D.R., Baranov V.I., Ornatsky O.I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J.E., Tanner S.D. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

