Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses
- PMID: 28475900
- PMCID: PMC5737939
- DOI: 10.1016/j.cell.2017.04.014
Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses
Abstract
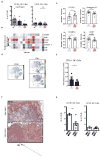
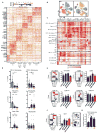
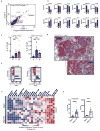
To guide the design of immunotherapy strategies for patients with early stage lung tumors, we developed a multiscale immune profiling strategy to map the immune landscape of early lung adenocarcinoma lesions to search for tumor-driven immune changes. Utilizing a barcoding method that allows a simultaneous single-cell analysis of the tumor, non-involved lung, and blood cells, we provide a detailed immune cell atlas of early lung tumors. We show that stage I lung adenocarcinoma lesions already harbor significantly altered T cell and NK cell compartments. Moreover, we identified changes in tumor-infiltrating myeloid cell (TIM) subsets that likely compromise anti-tumor T cell immunity. Paired single-cell analyses thus offer valuable knowledge of tumor-driven immune changes, providing a powerful tool for the rational design of immune therapies. VIDEO ABSTRACT.
Keywords: CD141+ DC; CD1c+ DC; NK Cell; T Cell; TIM; TLS; human non-small cell lung cancer (NSCLC); immune cell atlas; lung adenocarcinoma; tumor macrophage.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Heavy Metal Enlightens Tumor Immunity.Cell. 2017 May 4;169(4):567-569. doi: 10.1016/j.cell.2017.04.017. Cell. 2017. PMID: 28475889 Free PMC article.
-
Cell maps reveal fresh details on how the immune system fights cancer.Nature. 2017 May 4;545(7653):143. doi: 10.1038/nature.2017.21931. Nature. 2017. PMID: 28492266 No abstract available.
-
Tumour immunology: Immune atlas sheds light on anticancer responses.Nat Rev Immunol. 2017 May 26;17(6):347. doi: 10.1038/nri.2017.59. Nat Rev Immunol. 2017. PMID: 28548134 No abstract available.
-
Heavy Metal to Rock the Immune Infiltrate.Trends Immunol. 2017 Aug;38(8):539-541. doi: 10.1016/j.it.2017.05.007. Epub 2017 Jun 9. Trends Immunol. 2017. PMID: 28602618
-
Unmassked: Single-Cell Profiling of Immune Cell Populations in Tumors.Mol Ther. 2017 Aug 2;25(8):1745-1747. doi: 10.1016/j.ymthe.2017.06.024. Epub 2017 Jul 4. Mol Ther. 2017. PMID: 28684257 Free PMC article. No abstract available.
References
-
- Besse B, Johnson M, Janne PA, Janne PA, Garassino MC, Eberhardt WE, Peters S, Toh CK, Kurata T, Li Z, et al. 16LBA Phase II, single-arm trial (BIRCH) of atezolizumab as first-line or subsequent therapy for locally advanced or metastatic PD-L1-selected non-small cell lung cancer (NSCLC) European Journal of Cancer. 2015;51
-
- Black WC, Keeler EB, Soneji SS. Cost-effectiveness of CT screening in the National Lung Screening Trial. N Engl J Med. 2015;372:388. - PubMed
-
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of statistical mechanics: theory and experiment. 2008;2008:P10008.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

