iRNAm5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition
- PMID: 28476023
- PMCID: PMC5522291
- DOI: 10.18632/oncotarget.17104
iRNAm5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition
Abstract
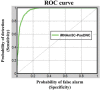
Occurring at cytosine (C) of RNA, 5-methylcytosine (m5C) is an important post-transcriptional modification (PTCM). The modification plays significant roles in biological processes by regulating RNA metabolism in both eukaryotes and prokaryotes. It may also, however, cause cancers and other major diseases. Given an uncharacterized RNA sequence that contains many C residues, can we identify which one of them can be of m5C modification, and which one cannot? It is no doubt a crucial problem, particularly with the explosive growth of RNA sequences in the postgenomic age. Unfortunately, so far no user-friendly web-server whatsoever has been developed to address such a problem. To meet the increasingly high demand from most experimental scientists working in the area of drug development, we have developed a new predictor called iRNAm5C-PseDNC by incorporating ten types of physical-chemical properties into pseudo dinucleotide composition via the auto/cross-covariance approach. Rigorous jackknife tests show that its anticipated accuracy is quite high. For most experimental scientists' convenience, a user-friendly web-server for the predictor has been provided at http://www.jci-bioinfo.cn/iRNAm5C-PseDNC along with a step-by-step user guide, by which users can easily obtain their desired results without the need to go through the complicated mathematical equations involved. It has not escaped our notice that the approach presented here can also be used to deal with many other problems in genome analysis.
Keywords: RNA 5-methylcytosine sites; auto/cross-covariance; physical-chemical property matrix; pseudo dinucleotide composition; web-server.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Gu XR, Gustafsson C, Ku J, Yu M, Santi DV. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:4053–4057. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

