Multi-omics approaches to disease
- PMID: 28476144
- PMCID: PMC5418815
- DOI: 10.1186/s13059-017-1215-1
Multi-omics approaches to disease
Abstract
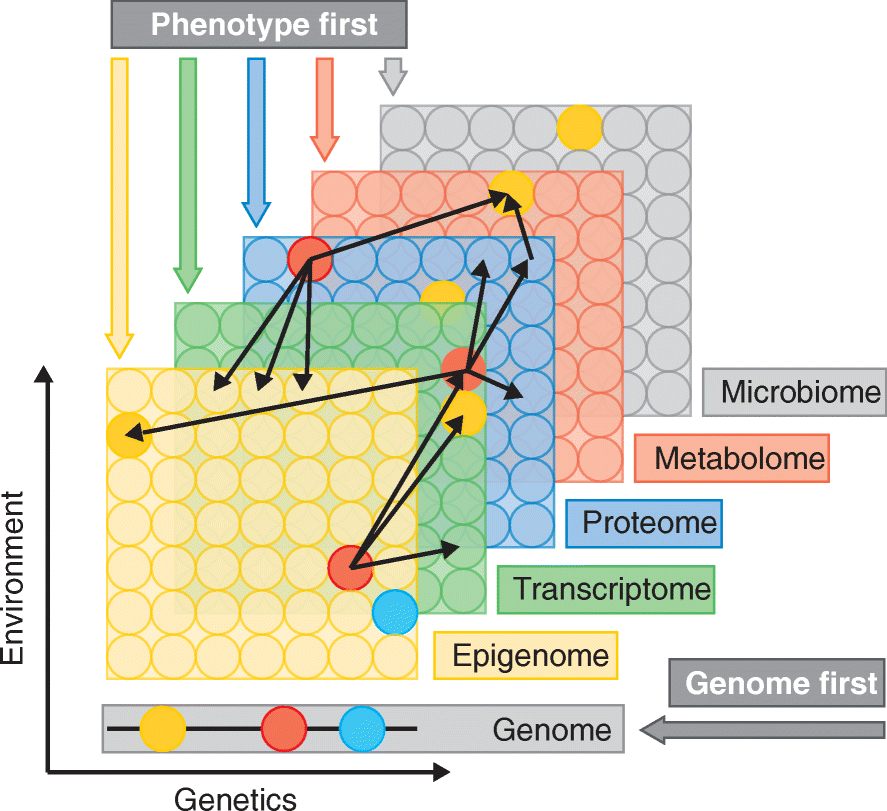
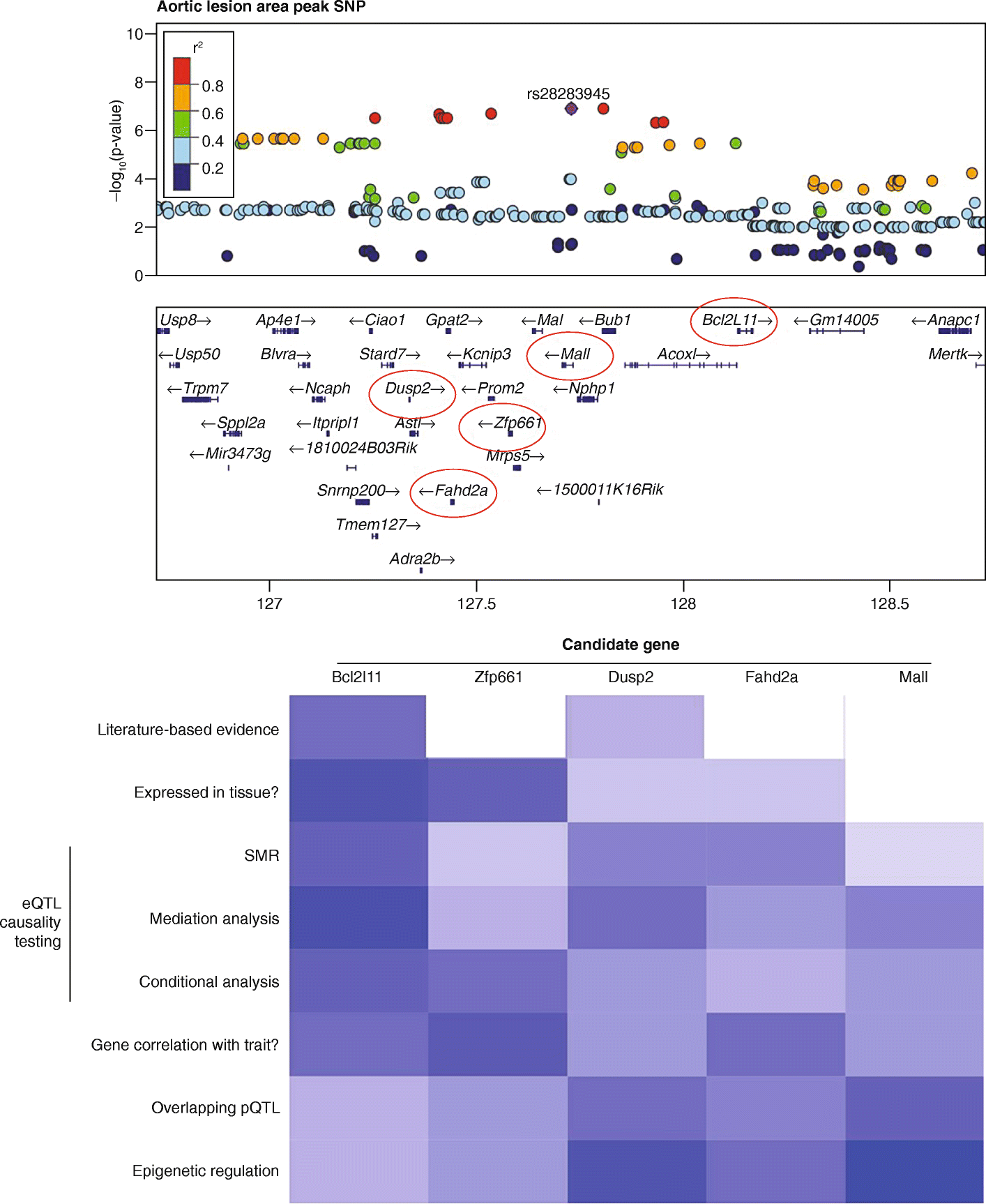
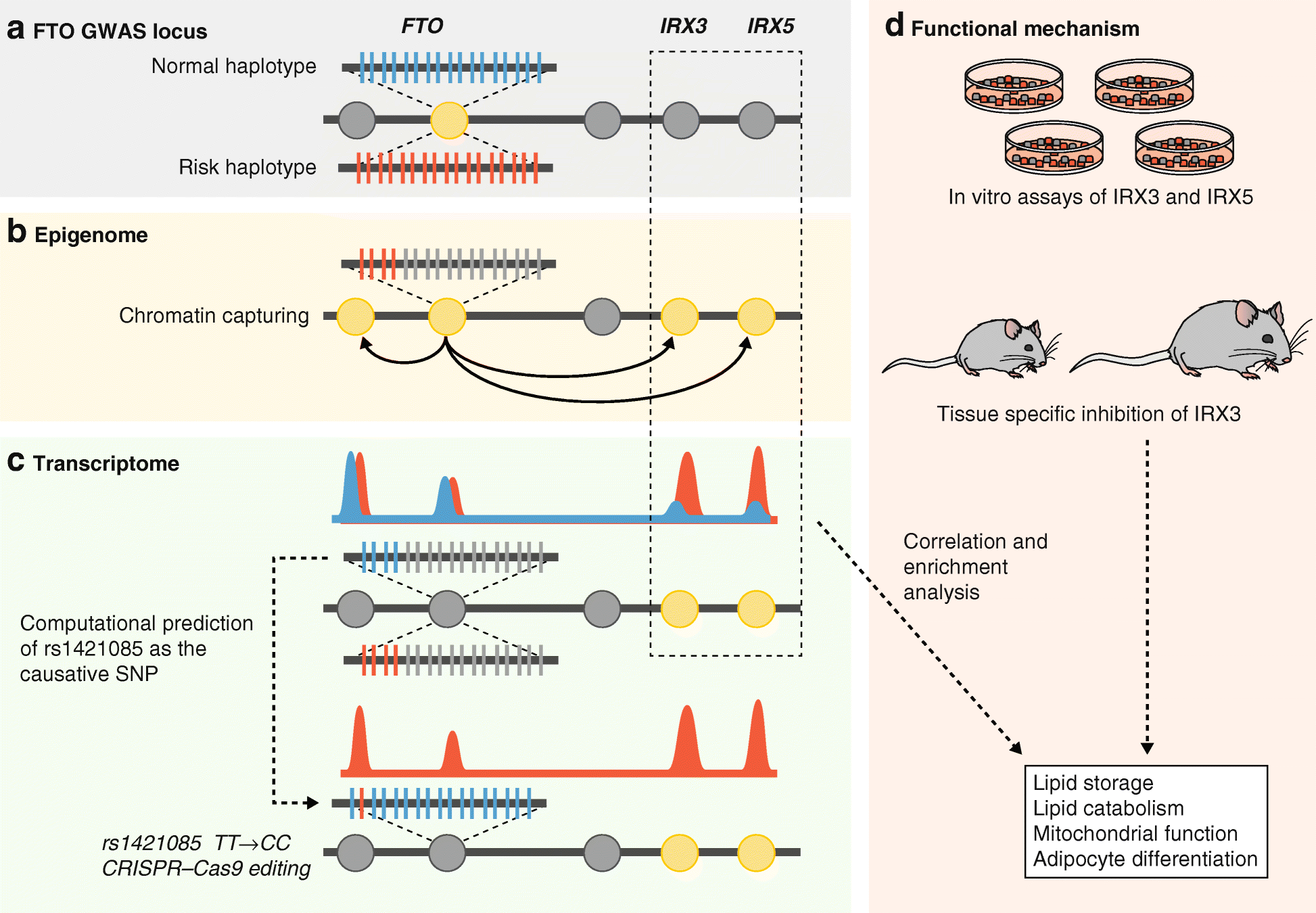
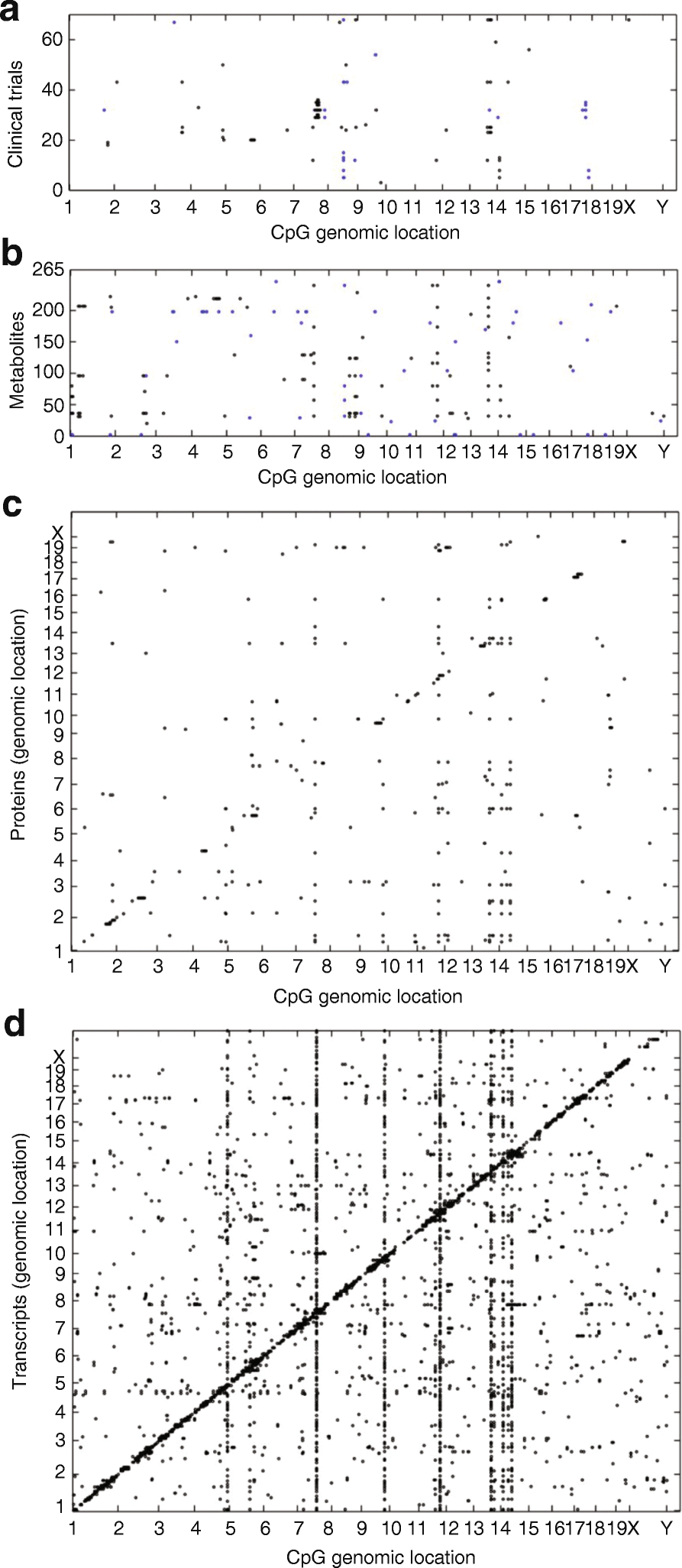
High-throughput technologies have revolutionized medical research. The advent of genotyping arrays enabled large-scale genome-wide association studies and methods for examining global transcript levels, which gave rise to the field of "integrative genetics". Other omics technologies, such as proteomics and metabolomics, are now often incorporated into the everyday methodology of biological researchers. In this review, we provide an overview of such omics technologies and focus on methods for their integration across multiple omics layers. As compared to studies of a single omics type, multi-omics offers the opportunity to understand the flow of information that underlies disease.
Figures
References
-
- Gibson G. A primer of human genetics. 1. Sunderland (Massachusetts): Sinauer Associates, Inc.; 2015.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

