Defining Genetic Fitness Determinants and Creating Genomic Resources for an Oral Pathogen
- PMID: 28476775
- PMCID: PMC5494627
- DOI: 10.1128/AEM.00797-17
Defining Genetic Fitness Determinants and Creating Genomic Resources for an Oral Pathogen
Abstract
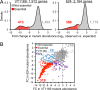
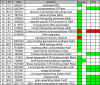
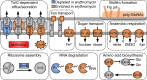
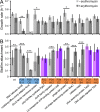
Periodontitis is a microbial infection that destroys the structures that support the teeth. Although it is typically a chronic condition, rapidly progressing, aggressive forms are associated with the oral pathogen Aggregatibacter actinomycetemcomitans One of this bacterium's key virulence traits is its ability to attach to surfaces and form robust biofilms that resist killing by the host and antibiotics. Though much has been learned about A. actinomycetemcomitans since its initial discovery, we lack insight into a fundamental aspect of its basic biology, as we do not know the full set of genes that it requires for viability (the essential genome). Furthermore, research on A. actinomycetemcomitans is hampered by the field's lack of a mutant collection. To address these gaps, we used rapid transposon mutant sequencing (Tn-seq) to define the essential genomes of two strains of A. actinomycetemcomitans, revealing a core set of 319 genes. We then generated an arrayed mutant library comprising >1,500 unique insertions and used a sequencing-based approach to define each mutant's position (well and plate) in the library. To demonstrate its utility, we screened the library for mutants with weakened resistance to subinhibitory erythromycin, revealing the multidrug efflux pump AcrAB as a critical resistance factor. During the screen, we discovered that erythromycin induces A. actinomycetemcomitans to form biofilms. We therefore devised a novel Tn-seq-based screen to identify specific factors that mediate this phenotype and in follow-up experiments confirmed 4 mutants. Together, these studies present new insights and resources for investigating the basic biology and disease mechanisms of a human pathogen.IMPORTANCE Millions suffer from gum disease, which often is caused by Aggregatibacter actinomycetemcomitans, a bacterium that forms antibiotic-resistant biofilms. To fully understand any organism, we should be able to answer: what genes does it require for life? Here, we address this question for A. actinomycetemcomitans by determining the genes in its genome that cannot be mutated. As for the genes that can be mutated, we archived these mutants into a library, which we used to find genes that contribute to antibiotic resistance, leading us to discover that antibiotics cause A. actinomycetemcomitans to form biofilms. We then devised an approach to find genes that mediate this process and confirmed 4 genes. These results illuminate new fundamental traits of a human pathogen.
Keywords: Aggregatibacter; Tn-seq; antibiotics; biofilms; essential genome.
Copyright © 2017 American Society for Microbiology.
Figures

Similar articles
-
The cagE gene sequence as a diagnostic marker to identify JP2 and non-JP2 highly leukotoxic Aggregatibacter actinomycetemcomitans serotype b strains.J Periodontal Res. 2017 Oct;52(5):903-912. doi: 10.1111/jre.12462. Epub 2017 Apr 10. J Periodontal Res. 2017. PMID: 28397250
-
Evolutionary Divergence of Aggregatibacter actinomycetemcomitans.J Dent Res. 2016 Jan;95(1):94-101. doi: 10.1177/0022034515608163. Epub 2015 Sep 29. J Dent Res. 2016. PMID: 26420795 Free PMC article.
-
Multilocus Sequence Typing of Aggregatibacter actinomycetemcomitans Competently Depicts the Population Structure of the Species.Microbiol Spectr. 2021 Dec 22;9(3):e0108521. doi: 10.1128/Spectrum.01085-21. Epub 2021 Dec 15. Microbiol Spectr. 2021. PMID: 34908433 Free PMC article.
-
From Global to Nano: A Geographical Perspective of Aggregatibacter actinomycetemcomitans.Pathogens. 2024 Sep 27;13(10):837. doi: 10.3390/pathogens13100837. Pathogens. 2024. PMID: 39452709 Free PMC article. Review.
-
Aggregatibacter actinomycetemcomitans: virulence of its leukotoxin and association with aggressive periodontitis.Virulence. 2015;6(3):188-95. doi: 10.4161/21505594.2014.982428. Virulence. 2015. PMID: 25494963 Free PMC article. Review.
Cited by
-
Elucidation of Essential Genes and Mutant Fitness during Adaptation toward Nitrogen Fixation Conditions in the Endophyte Azoarcus olearius BH72 Revealed by Tn-Seq.Microbiol Spectr. 2022 Dec 21;10(6):e0216222. doi: 10.1128/spectrum.02162-22. Epub 2022 Nov 23. Microbiol Spectr. 2022. PMID: 36416558 Free PMC article.
-
Genome-Wide Investigation of Pasteurella multocida Identifies the Stringent Response as a Negative Regulator of Hyaluronic Acid Capsule Production.Microbiol Spectr. 2022 Apr 27;10(2):e0019522. doi: 10.1128/spectrum.00195-22. Epub 2022 Apr 11. Microbiol Spectr. 2022. PMID: 35404102 Free PMC article.
-
Aggregatibacter actinomycetemcomitans colonization and persistence in a primate model.Proc Natl Acad Sci U S A. 2019 Oct 29;116(44):22307-22313. doi: 10.1073/pnas.1905238116. Epub 2019 Oct 14. Proc Natl Acad Sci U S A. 2019. PMID: 31611409 Free PMC article.
-
Dental Plaque Microbial Resistomes of Periodontal Health and Disease and Their Changes after Scaling and Root Planing Therapy.mSphere. 2021 Aug 25;6(4):e0016221. doi: 10.1128/mSphere.00162-21. Epub 2021 Jul 21. mSphere. 2021. PMID: 34287005 Free PMC article.
-
Identification of essential genes in Coxiella burnetii.Microb Genom. 2023 Feb;9(2):mgen000944. doi: 10.1099/mgen.0.000944. Microb Genom. 2023. PMID: 36723494 Free PMC article.
References
-
- Fine DH, Markowitz K, Fairlie K, Tischio-Bereski D, Ferrendiz J, Furgang D, Paster BJ, Dewhirst FE. 2013. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol 51:2850–2861. doi:10.1128/JCM.00729-13. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

