An Evolutionary Perspective on Yeast Mating-Type Switching
- PMID: 28476860
- PMCID: PMC5419495
- DOI: 10.1534/genetics.117.202036
An Evolutionary Perspective on Yeast Mating-Type Switching
Abstract
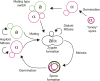
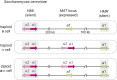
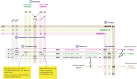
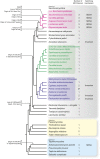
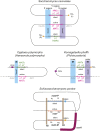
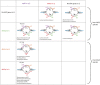
Cell differentiation in yeast species is controlled by a reversible, programmed DNA-rearrangement process called mating-type switching. Switching is achieved by two functionally similar but structurally distinct processes in the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe In both species, haploid cells possess one active and two silent copies of the mating-type locus (a three-cassette structure), the active locus is cleaved, and synthesis-dependent strand annealing is used to replace it with a copy of a silent locus encoding the opposite mating-type information. Each species has its own set of components responsible for regulating these processes. In this review, we summarize knowledge about the function and evolution of mating-type switching components in these species, including mechanisms of heterochromatin formation, MAT locus cleavage, donor bias, lineage tracking, and environmental regulation of switching. We compare switching in these well-studied species to others such as Kluyveromyces lactis and the methylotrophic yeasts Ogataea polymorpha and Komagataella phaffii We focus on some key questions: Which cells switch mating type? What molecular apparatus is required for switching? Where did it come from? And what is the evolutionary purpose of switching?
Keywords: evolution; homothallism; mating-type switching; sporulation; yeast genetics.
Copyright © 2017 by the Genetics Society of America.
Figures
Similar articles
-
Flip/flop mating-type switching in the methylotrophic yeast Ogataea polymorpha is regulated by an Efg1-Rme1-Ste12 pathway.PLoS Genet. 2017 Nov 27;13(11):e1007092. doi: 10.1371/journal.pgen.1007092. eCollection 2017 Nov. PLoS Genet. 2017. PMID: 29176810 Free PMC article.
-
Regulation of mating type switching by the mating type genes and RME1 in Ogataea polymorpha.Sci Rep. 2017 Nov 24;7(1):16318. doi: 10.1038/s41598-017-16284-7. Sci Rep. 2017. PMID: 29176579 Free PMC article.
-
Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha.PLoS Genet. 2014 Nov 20;10(11):e1004796. doi: 10.1371/journal.pgen.1004796. eCollection 2014 Nov. PLoS Genet. 2014. PMID: 25412462 Free PMC article.
-
Mating-Type Switching in Budding Yeasts, from Flip/Flop Inversion to Cassette Mechanisms.Microbiol Mol Biol Rev. 2022 Jun 15;86(2):e0000721. doi: 10.1128/mmbr.00007-21. Epub 2022 Feb 23. Microbiol Mol Biol Rev. 2022. PMID: 35195440 Free PMC article. Review.
-
Mating-type genes and MAT switching in Saccharomyces cerevisiae.Genetics. 2012 May;191(1):33-64. doi: 10.1534/genetics.111.134577. Genetics. 2012. PMID: 22555442 Free PMC article. Review.
Cited by
-
The Ecology and Evolution of the Baker's Yeast Saccharomyces cerevisiae.Genes (Basel). 2022 Jan 26;13(2):230. doi: 10.3390/genes13020230. Genes (Basel). 2022. PMID: 35205274 Free PMC article. Review.
-
Shuttling Homeoproteins and Their Biological Significance.Methods Mol Biol. 2022;2383:33-44. doi: 10.1007/978-1-0716-1752-6_2. Methods Mol Biol. 2022. PMID: 34766280 Review.
-
Mitochondrial Inheritance in Phytopathogenic Fungi-Everything Is Known, or Is It?Int J Mol Sci. 2020 May 29;21(11):3883. doi: 10.3390/ijms21113883. Int J Mol Sci. 2020. PMID: 32485941 Free PMC article. Review.
-
Pneumocystis Mating-Type Locus and Sexual Cycle during Infection.Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0000921. doi: 10.1128/MMBR.00009-21. Epub 2021 Jun 16. Microbiol Mol Biol Rev. 2021. PMID: 34132101 Free PMC article. Review.
-
Highly Diverse Phytophthora infestans Populations Infecting Potato Crops in Pskov Region, North-West Russia.J Fungi (Basel). 2022 Apr 30;8(5):472. doi: 10.3390/jof8050472. J Fungi (Basel). 2022. PMID: 35628727 Free PMC article.
References
-
- Arcangioli B., Thon G., 2004. Mating type cassettes: structure, switching and silencing, The Molecular Biology of Schizosaccharomyces Pombe, edited by Egel R. Springer-Verlag, Berlin.
-
- Astell C. R., Ahlstrom-Jonasson L., Smith M., Tatchell K., Nasmyth K. A., et al. , 1981. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell 27: 15–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

