Insights into the catalysis of a lysine-tryptophan bond in bacterial peptides by a SPASM domain radical S-adenosylmethionine (SAM) peptide cyclase
- PMID: 28476884
- PMCID: PMC5491770
- DOI: 10.1074/jbc.M117.783464
Insights into the catalysis of a lysine-tryptophan bond in bacterial peptides by a SPASM domain radical S-adenosylmethionine (SAM) peptide cyclase
Abstract
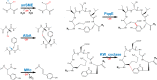
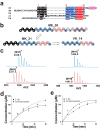
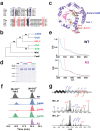
Radical S-adenosylmethionine (SAM) enzymes are emerging as a major superfamily of biological catalysts involved in the biosynthesis of the broad family of bioactive peptides called ribosomally synthesized and post-translationally modified peptides (RiPPs). These enzymes have been shown to catalyze unconventional reactions, such as methyl transfer to electrophilic carbon atoms, sulfur to Cα atom thioether bonds, or carbon-carbon bond formation. Recently, a novel radical SAM enzyme catalyzing the formation of a lysine-tryptophan bond has been identified in Streptococcus thermophilus, and a reaction mechanism has been proposed. By combining site-directed mutagenesis, biochemical assays, and spectroscopic analyses, we show here that this enzyme, belonging to the emerging family of SPASM domain radical SAM enzymes, likely contains three [4Fe-4S] clusters. Notably, our data support that the seven conserved cysteine residues, present within the SPASM domain, are critical for enzyme activity. In addition, we uncovered the minimum substrate requirements and demonstrate that KW cyclic peptides are more widespread than anticipated, notably in pathogenic bacteria. Finally, we show a strict specificity of the enzyme for lysine and tryptophan residues and the dependence of an eight-amino acid leader peptide for activity. Altogether, our study suggests novel mechanistic links among SPASM domain radical SAM enzymes and supports the involvement of non-cysteinyl ligands in the coordination of auxiliary clusters.
Keywords: biosynthesis; enzyme catalysis; enzyme mechanism; iron-sulfur protein; metalloenzyme; peptide biosynthesis; radical; radical AdoMet; radical SAM; radical SAM enzyme.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Mehta A. P., Abdelwahed S. H., Mahanta N., Fedoseyenko D., Philmus B., Cooper L. E., Liu Y., Jhulki I., Ealick S. E., and Begley T. P. (2015) Radical S-adenosylmethionine (SAM) enzymes in cofactor biosynthesis: a treasure trove of complex organic radical rearrangement reactions. J. Biol. Chem. 290, 3980–3986 - PMC - PubMed
-
- Benjdia A., and Berteau O. (2016) Sulfatases and radical SAM enzymes: emerging themes in glycosaminoglycan metabolism and the human microbiota. Biochem. Soc. Trans. 44, 109–115 - PubMed
-
- Berteau O., and Benjdia A. (2017) DNA repair by the radical SAM enzyme spore photoproduct lyase: from biochemistry to structural investigations. Photochem. Photobiol. 93, 67–77 - PubMed
-
- Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., et al. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

