Caught in the act - protein adaptation and the expanding roles of the PACS proteins in tissue homeostasis and disease
- PMID: 28476937
- PMCID: PMC5482974
- DOI: 10.1242/jcs.199463
Caught in the act - protein adaptation and the expanding roles of the PACS proteins in tissue homeostasis and disease
Abstract
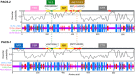
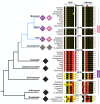
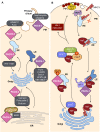
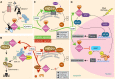
Vertebrate proteins that fulfill multiple and seemingly disparate functions are increasingly recognized as vital solutions to maintaining homeostasis in the face of the complex cell and tissue physiology of higher metazoans. However, the molecular adaptations that underpin this increased functionality remain elusive. In this Commentary, we review the PACS proteins - which first appeared in lower metazoans as protein traffic modulators and evolved in vertebrates to integrate cytoplasmic protein traffic and interorganellar communication with nuclear gene expression - as examples of protein adaptation 'caught in the act'. Vertebrate PACS-1 and PACS-2 increased their functional density and roles as metabolic switches by acquiring phosphorylation sites and nuclear trafficking signals within disordered regions of the proteins. These findings illustrate one mechanism by which vertebrates accommodate their complex cell physiology with a limited set of proteins. We will also highlight how pathogenic viruses exploit the PACS sorting pathways as well as recent studies on PACS genes with mutations or altered expression that result in diverse diseases. These discoveries suggest that investigation of the evolving PACS protein family provides a rich opportunity for insight into vertebrate cell and organ homeostasis.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
Similar articles
-
Akt and 14-3-3 control a PACS-2 homeostatic switch that integrates membrane traffic with TRAIL-induced apoptosis.Mol Cell. 2009 May 14;34(4):497-509. doi: 10.1016/j.molcel.2009.04.011. Mol Cell. 2009. PMID: 19481529 Free PMC article.
-
At the crossroads of homoeostasis and disease: roles of the PACS proteins in membrane traffic and apoptosis.Biochem J. 2009 Jun 12;421(1):1-15. doi: 10.1042/BJ20081016. Biochem J. 2009. PMID: 19505291 Free PMC article. Review.
-
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) protein-induced lysosomal translocation of proapoptotic effectors is mediated by phosphofurin acidic cluster sorting protein-2 (PACS-2).J Biol Chem. 2012 Jul 13;287(29):24427-37. doi: 10.1074/jbc.M112.342238. Epub 2012 May 29. J Biol Chem. 2012. PMID: 22645134 Free PMC article.
-
Roles of FAM134B in diseases from the perspectives of organelle membrane morphogenesis and cellular homeostasis.J Cell Physiol. 2021 Oct;236(10):7242-7255. doi: 10.1002/jcp.30377. Epub 2021 Apr 12. J Cell Physiol. 2021. PMID: 33843059 Review.
-
Cellular inhibitor of apoptosis (cIAP)-mediated ubiquitination of phosphofurin acidic cluster sorting protein 2 (PACS-2) negatively regulates tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) cytotoxicity.PLoS One. 2014 Mar 14;9(3):e92124. doi: 10.1371/journal.pone.0092124. eCollection 2014. PLoS One. 2014. PMID: 24633224 Free PMC article.
Cited by
-
Multiomic Predictors of Short-Term Weight Loss and Clinical Outcomes During a Behavioral-Based Weight Loss Intervention.Obesity (Silver Spring). 2021 May;29(5):859-869. doi: 10.1002/oby.23127. Epub 2021 Apr 3. Obesity (Silver Spring). 2021. PMID: 33811477 Free PMC article.
-
Prognostic role of alternative splicing events in head and neck squamous cell carcinoma.Cancer Cell Int. 2020 May 14;20:168. doi: 10.1186/s12935-020-01249-0. eCollection 2020. Cancer Cell Int. 2020. PMID: 32467664 Free PMC article.
-
Calcium flux control by Pacs1-Wdr37 promotes lymphocyte quiescence and lymphoproliferative diseases.EMBO J. 2021 May 3;40(9):e104888. doi: 10.15252/embj.2020104888. Epub 2021 Feb 25. EMBO J. 2021. PMID: 33630350 Free PMC article.
-
PACS2, PACS1, and VACTERL: A Clinical Overlap.Mol Syndromol. 2025 Feb;16(1):29-32. doi: 10.1159/000539473. Epub 2024 Aug 7. Mol Syndromol. 2025. PMID: 39911171 Free PMC article.
-
Exploring prognostic markers for patients with acute myeloid leukemia based on cuproptosis related genes.Transl Cancer Res. 2023 Aug 31;12(8):2008-2022. doi: 10.21037/tcr-23-85. Epub 2023 Aug 28. Transl Cancer Res. 2023. PMID: 37701119 Free PMC article.
References
-
- Anderson G. R., Brenner B. M., Swede H., Chen N., Henry W. M., Conroy J. M., Karpenko M. J., Issa J. P., Bartos J. D., Brunelle J.K. et al. (2001). Intrachromosomal genomic instability in human sporadic colorectal cancer measured by genome-wide allelotyping and inter-(simple sequence repeat) PCR. Cancer Res. 61, 8274-8283. - PubMed
-
- Area-Gomez E., del Carmen Lara Castillo M., Tambini M. D., Guardia-Laguarta C., de Groof A. J. C., Madra M., Ikenouchi J., Umeda M., Bird T. D., Sturley S. L. et al. (2012). Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31, 4106-4123. 10.1038/emboj.2012.202 - DOI - PMC - PubMed
-
- Arnoult D., Bartle L. M., Skaletskaya A., Poncet D., Zamzami N., Park P. U., Sharpe J., Youle R. J. and Goldmacher V. S. (2004). Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. USA 101, 7988-7993. 10.1073/pnas.0401897101 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

