Comparative molecular dynamics study of neuromyelitis optica-immunoglobulin G binding to aquaporin-4 extracellular domains
- PMID: 28477975
- PMCID: PMC5581535
- DOI: 10.1016/j.bbamem.2017.05.001
Comparative molecular dynamics study of neuromyelitis optica-immunoglobulin G binding to aquaporin-4 extracellular domains
Abstract
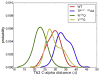
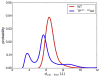
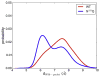
Neuromyelitis optica (NMO) is an inflammatory demyelinating disease of the central nervous system in which most patients have serum autoantibodies (called NMO-IgG) that bind to astrocyte water channel aquaporin-4 (AQP4). A potential therapeutic strategy in NMO is to block the interaction of NMO-IgG with AQP4. Building on recent observation that some single-point and compound mutations of the AQP4 extracellular loop C prevent NMO-IgG binding, we carried out comparative Molecular Dynamics (MD) investigations on three AQP4 mutants, TP137-138AA, N153Q and V150G, whose 295-ns long trajectories were compared to that of wild type human AQP4. A robust conclusion of our modeling is that loop C mutations affect the conformation of neighboring extracellular loop A, thereby interfering with NMO-IgG binding. Analysis of individual mutations suggested specific hydrogen bonding and other molecular interactions involved in AQP4-IgG binding to AQP4.
Keywords: Aquaporin-4; Molecular Dynamics; Mutations; Neuromyelitis Optica.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures
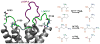




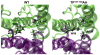
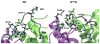
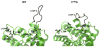

References
-
- Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem. 1999;68:425–458. - PubMed
-
- Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. - PubMed
-
- Sachdeva R, Singh B. Insights into structural mechanisms of gating induced regulation of aquaporins. Prog Biophys Mol Biol. 2014;114:69–79. - PubMed
-
- Ilan B, Tajkhorshid E, Schulten K, Voth GA. The mechanism of proton exclusion in aquaporin channels. Proteins Struct Funct Bioinforma. 2004;55:223–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

