Re: Genome-Wide Association Study Identifies African-Specific Susceptibility Loci in African Americans With Inflammatory Bowel Disease
- PMID: 28478146
- PMCID: PMC6033331
- DOI: 10.1053/j.gastro.2017.02.041
Re: Genome-Wide Association Study Identifies African-Specific Susceptibility Loci in African Americans With Inflammatory Bowel Disease
Abstract
BACKGROUND & AIMS: The inflammatory bowel diseases (IBD) ulcerative colitis (UC) and Crohn’s disease (CD) cause significant morbidity and are increasing in prevalence among all populations, including African Americans. More than 200 susceptibility loci have been identified in populations of predominantly European ancestry, but few loci have been associated with IBD in other ethnicities.
METHODS: We performed 2 high-density, genome-wide scans comprising 2345 cases of African Americans with IBD (1646 with CD, 583 with UC, and 116 inflammatory bowel disease unclassified) and 5002 individuals without IBD (controls, identified from the Health Retirement Study and Kaiser Permanente database). Single-nucleotide polymorphisms (SNPs) associated at P < 5.0 × 10−8 in meta-analysis with a nominal evidence (P < .05) in each scan were considered to have genome-wide significance.
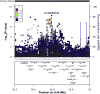
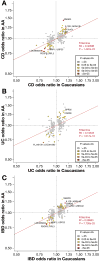
RESULTS: We detected SNPs at HLA-DRB1, and African-specific SNPs at ZNF649 and LSAMP, with associations of genome-wide significance for UC. We detected SNPs at USP25 with associations of genome-wide significance for IBD. No associations of genome-wide significance were detected for CD. In addition, 9 genes previously associated with IBD contained SNPs with significant evidence for replication (P < 1.6 × 10−6): ADCY3, CXCR6, HLA-DRB1 to HLA-DQA1 (genome-wide signifi-cance on conditioning), IL12B, PTGER4, and TNC for IBD; IL23R, PTGER4, and SNX20 (in strong linkage disequilibrium with NOD2) for CD; and KCNQ2 (near TNFRSF6B) for UC. Several of these genes, such as TNC (near TNFSF15), CXCR6, and genes associated with IBD at the HLA locus, contained SNPs with unique association patterns with African-specific alleles.
CONCLUSIONS: We performed a genome-wide association study of African Americans with IBD and identified loci associated with UC in only this population; we also replicated IBD, CD, and UC loci identified in European populations. The detection of variants associated with IBD risk in only people of African descent demonstrates the importance of studying the genetics of IBD and other complex diseases in populations beyond those of European ancestry.
Conflict of interest statement
The authors disclose no conflicts.
Figures


Comment in
-
Reply.Gastroenterology. 2017 Jun;152(8):2083-2084. doi: 10.1053/j.gastro.2017.05.006. Epub 2017 May 5. Gastroenterology. 2017. PMID: 28482170 Free PMC article. No abstract available.
Comment on
-
Genome-Wide Association Study Identifies African-Specific Susceptibility Loci in African Americans With Inflammatory Bowel Disease.Gastroenterology. 2017 Jan;152(1):206-217.e2. doi: 10.1053/j.gastro.2016.09.032. Epub 2016 Sep 28. Gastroenterology. 2017. PMID: 27693347 Free PMC article.
References
-
- Juyal G, Negi S, Sood A, et al. Genome-wide association scan in north Indians reveals three novel HLA-independent risk loci for ulcerative colitis. Gut. 2015;64:571–579. - PubMed
Publication types
MeSH terms
Grants and funding
- RC4 AG039029/AG/NIA NIH HHS/United States
- U54 DE023789/DE/NIDCR NIH HHS/United States
- U01 DK062413/DK/NIDDK NIH HHS/United States
- U01 AG009740/AG/NIA NIH HHS/United States
- U24 DK062429/DK/NIDDK NIH HHS/United States
- U01 DK062432/DK/NIDDK NIH HHS/United States
- U01 DK062429/DK/NIDDK NIH HHS/United States
- U01 DK062422/DK/NIDDK NIH HHS/United States
- RC2 AG036495/AG/NIA NIH HHS/United States
- U01 DK062423/DK/NIDDK NIH HHS/United States
- U01 AI067068/AI/NIAID NIH HHS/United States
- P01 DK046763/DK/NIDDK NIH HHS/United States
- U01 DK062420/DK/NIDDK NIH HHS/United States
- U01 DK062431/DK/NIDDK NIH HHS/United States
- R01 DK087694/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

