FRET studies of various conformational states adopted by transthyretin
- PMID: 28478513
- PMCID: PMC11107560
- DOI: 10.1007/s00018-017-2533-x
FRET studies of various conformational states adopted by transthyretin
Abstract
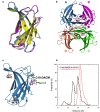
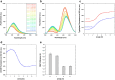
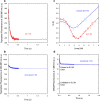
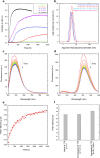
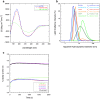
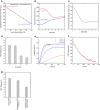
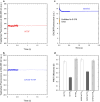
Transthyretin (TTR) is an extracellular protein able to deposit into well-defined protein aggregates called amyloid, in pathological conditions known as senile systemic amyloidosis, familial amyloid polyneuropathy, familial amyloid cardiomyopathy and leptomeningeal amyloidosis. At least three distinct partially folded states have been described for TTR, including the widely studied amyloidogenic state at mildly acidic pH. Here, we have used fluorescence resonance energy transfer (FRET) experiments in a monomeric variant of TTR (M-TTR) and in its W41F and W79F mutants, taking advantage of the presence of a unique, solvent-exposed, cysteine residue at position 10, that we have labelled with a coumarin derivative (DACM, acceptor), and of the two natural tryptophan residues at positions 41 and 79 (donors). Trp41 is located in an ideal position as it is one of the residues of β-strand C, whose degree of unfolding is debated. We found that the amyloidogenic state at low pH has the same FRET efficiency as the folded state at neutral pH in both M-TTR and W79F-M-TTR, indicating an unmodified Cys10-Trp41 distance. The partially folded state populated at low denaturant concentrations also has a similar FRET efficiency, but other spectroscopic probes indicate that it is distinct from the amyloidogenic state at acidic pH. By contrast, the off-pathway state accumulating transiently during refolding has a higher FRET efficiency, indicating non-native interactions that reduce the Cys10-Trp41 spatial distance, revealing a third distinct conformational state. Overall, our results clarify a negligible degree of unfolding of β-strand C in the formation of the amyloidogenic state and establish the concept that TTR is a highly plastic protein able to populate at least three distinct conformational states.
Keywords: FAC; FAP; Folding intermediate; Protein aggregation; Protein folding; Protein misfolding; SSA.
Figures


References
-
- Soprano DR, Herbert J, Soprano KJ, Schon EA, Goodman DS. Demonstration of transthyretin mRNA in the brain and other extrahepatic tissues in the rat. J Biol Chem. 1985;260(21):11793–11798. - PubMed
-
- Herbert J, Wilcox JN, Pham KT, Fremeau RT, Jr, Zeviani M, Dwork A, Soprano DR, Makover A, Goodman DS, Zimmerman EA, et al. Transthyretin: a choroid plexus-specific transport protein in human brain. The 1986 S. Weir Mitchell award. Neurology. 1986;36(7):900–911. doi: 10.1212/WNL.36.7.900. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

