A One-Step PCR-Based Assay to Evaluate the Efficiency and Precision of Genomic DNA-Editing Tools
- PMID: 28480303
- PMCID: PMC5415314
- DOI: 10.1016/j.omtm.2017.03.001
A One-Step PCR-Based Assay to Evaluate the Efficiency and Precision of Genomic DNA-Editing Tools
Abstract
Despite rapid progress, many problems and limitations persist and limit the applicability of gene-editing techniques. Making use of meganucleases, TALENs, or CRISPR/Cas9-based tools requires an initial step of pre-screening to determine the efficiency and specificity of the designed tools. This step remains time consuming and material consuming. Here we propose a simple, cheap, reliable, time-saving, and highly sensitive method to evaluate a given gene-editing tool based on its capacity to induce chromosomal translocations when combined with a reference engineered nuclease. In the proposed technique, designated engineered nuclease-induced translocations (ENIT), a plasmid coding for the DNA-editing tool to be tested is co-transfected into carefully chosen target cells along with that for an engineered nuclease of known specificity and efficiency. If the new enzyme efficiently cuts within the desired region, then specific chromosomal translocations will be generated between the two targeted genomic regions and be readily detectable by a one-step PCR or qPCR assay. The PCR product thus obtained can be directly sequenced, thereby determining the exact position of the double-strand breaks induced by the gene-editing tools. As a proof of concept, ENIT was successfully tested in different cell types and with different meganucleases, TALENs, and CRISPR/Cas9-based editing tools.
Keywords: CRISPR/Cas9; PCR; TALEN; assay; translocations.
Figures
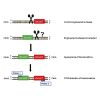
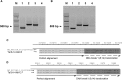



Similar articles
-
Comparison of CRISPR/Cas9 and TALENs on editing an integrated EGFP gene in the genome of HEK293FT cells.Springerplus. 2016 Jun 21;5(1):814. doi: 10.1186/s40064-016-2536-3. eCollection 2016. Springerplus. 2016. PMID: 27390654 Free PMC article.
-
Comparison of gene editing efficiencies of CRISPR/Cas9 and TALEN for generation of MSTN knock-out cashmere goats.Theriogenology. 2019 Jul 1;132:1-11. doi: 10.1016/j.theriogenology.2019.03.029. Epub 2019 Apr 1. Theriogenology. 2019. PMID: 30981084
-
Optimized CRISPR-Cas9 Genome Editing for Leishmania and Its Use To Target a Multigene Family, Induce Chromosomal Translocation, and Study DNA Break Repair Mechanisms.mSphere. 2017 Jan 18;2(1):e00340-16. doi: 10.1128/mSphere.00340-16. eCollection 2017 Jan-Feb. mSphere. 2017. PMID: 28124028 Free PMC article.
-
Gene Editing With TALEN and CRISPR/Cas in Rice.Prog Mol Biol Transl Sci. 2017;149:81-98. doi: 10.1016/bs.pmbts.2017.04.006. Epub 2017 May 24. Prog Mol Biol Transl Sci. 2017. PMID: 28712502 Review.
-
Applications of Alternative Nucleases in the Age of CRISPR/Cas9.Int J Mol Sci. 2017 Nov 29;18(12):2565. doi: 10.3390/ijms18122565. Int J Mol Sci. 2017. PMID: 29186020 Free PMC article. Review.
Cited by
-
Factors That Affect the Formation of Chromosomal Translocations in Cells.Cancers (Basel). 2022 Oct 18;14(20):5110. doi: 10.3390/cancers14205110. Cancers (Basel). 2022. PMID: 36291894 Free PMC article. Review.
-
HIV-1 Tat protein induces DNA damage in human peripheral blood B-lymphocytes via mitochondrial ROS production.Redox Biol. 2018 May;15:97-108. doi: 10.1016/j.redox.2017.11.024. Epub 2017 Dec 7. Redox Biol. 2018. PMID: 29220699 Free PMC article.
-
Ectopic Recruitment of the CTCF N-Terminal Domain with Two Proximal Zinc-Finger Domains as a Tool for 3D Genome Engineering.Int J Mol Sci. 2025 Aug 1;26(15):7446. doi: 10.3390/ijms26157446. Int J Mol Sci. 2025. PMID: 40806572 Free PMC article.
-
Specificity of cancer-related chromosomal translocations is linked to proximity after the DNA double-strand break and subsequent selection.NAR Cancer. 2023 Sep 23;5(3):zcad049. doi: 10.1093/narcan/zcad049. eCollection 2023 Sep. NAR Cancer. 2023. PMID: 37750169 Free PMC article.
-
LION: a simple and rapid method to achieve CRISPR gene editing.Cell Mol Life Sci. 2019 Jul;76(13):2633-2645. doi: 10.1007/s00018-019-03064-x. Epub 2019 Mar 18. Cell Mol Life Sci. 2019. PMID: 30887099 Free PMC article.
References
-
- Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. - PubMed
-
- Saada Y.B., Dib C., Lipinski M., Vassetzky Y.S. Genome- and cell-based strategies in therapy of muscular dystrophies. Biochemistry (Mosc.) 2016;81:678–690. - PubMed
-
- Hayashi K., Yandell D.W. How sensitive is PCR-SSCP? Hum. Mutat. 1993;2:338–346. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

