Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment
- PMID: 28480880
- PMCID: PMC5424149
- DOI: 10.1038/ncomms15134
Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment
Abstract
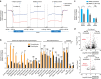
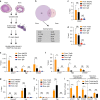
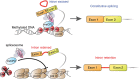
While intron retention (IR) is considered a widely conserved and distinct mechanism of gene expression control, its regulation is poorly understood. Here we show that DNA methylation directly regulates IR. We also find reduced occupancy of MeCP2 near the splice junctions of retained introns, mirroring the reduced DNA methylation at these sites. Accordingly, MeCP2 depletion in tissues and cells enhances IR. By analysing the MeCP2 interactome using mass spectrometry and RNA co-precipitation, we demonstrate that decreased MeCP2 binding near splice junctions facilitates IR via reduced recruitment of splicing factors, including Tra2b, and increased RNA polymerase II stalling. These results suggest an association between IR and a slower rate of transcription elongation, which reflects inefficient splicing factor recruitment. In summary, our results reinforce the interdependency between alternative splicing involving IR and epigenetic controls of gene expression.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
An important class of intron retention events in human erythroblasts is regulated by cryptic exons proposed to function as splicing decoys.RNA. 2018 Sep;24(9):1255-1265. doi: 10.1261/rna.066951.118. Epub 2018 Jun 29. RNA. 2018. PMID: 29959282 Free PMC article.
-
The alternative role of DNA methylation in splicing regulation.Trends Genet. 2015 May;31(5):274-80. doi: 10.1016/j.tig.2015.03.002. Epub 2015 Mar 30. Trends Genet. 2015. PMID: 25837375 Review.
-
Antisense targeting of decoy exons can reduce intron retention and increase protein expression in human erythroblasts.RNA. 2020 Aug;26(8):996-1005. doi: 10.1261/rna.075028.120. Epub 2020 Apr 20. RNA. 2020. PMID: 32312846 Free PMC article.
-
Regulation of mRNA splicing by MeCP2 via epigenetic modifications in the brain.Sci Rep. 2017 Feb 17;7:42790. doi: 10.1038/srep42790. Sci Rep. 2017. PMID: 28211484 Free PMC article.
-
Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):6800-6. doi: 10.1073/pnas.1411269112. Epub 2015 Mar 4. Proc Natl Acad Sci U S A. 2015. PMID: 25739960 Free PMC article. Review.
Cited by
-
Dissecting splicing decisions and cell-to-cell variability with designed sequence libraries.Nat Commun. 2019 Oct 8;10(1):4572. doi: 10.1038/s41467-019-12642-3. Nat Commun. 2019. PMID: 31594945 Free PMC article.
-
Switched alternative splicing events as attractive features in lung squamous cell carcinoma.Cancer Cell Int. 2022 Jan 5;22(1):5. doi: 10.1186/s12935-021-02429-2. Cancer Cell Int. 2022. PMID: 34986865 Free PMC article.
-
Population-dependent Intron Retention and DNA Methylation in Breast Cancer.Mol Cancer Res. 2018 Mar;16(3):461-469. doi: 10.1158/1541-7786.MCR-17-0227. Epub 2018 Jan 12. Mol Cancer Res. 2018. PMID: 29330282 Free PMC article.
-
The Crosstalk Between Epigenetic Mechanisms and Alternative RNA Processing Regulation.Front Genet. 2020 Aug 20;11:998. doi: 10.3389/fgene.2020.00998. eCollection 2020. Front Genet. 2020. PMID: 32973889 Free PMC article. Review.
-
Alternative splicing plays key roles in response to stress across different stages of fighting in the fish Betta splendens.BMC Genomics. 2022 May 30;22(Suppl 5):920. doi: 10.1186/s12864-022-08609-2. BMC Genomics. 2022. PMID: 35637454 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

