Nucleic Acid Templated Reactions for Chemical Biology
- PMID: 28480997
- PMCID: PMC5488204
- DOI: 10.1002/cmdc.201700266
Nucleic Acid Templated Reactions for Chemical Biology
Abstract
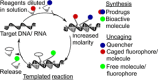
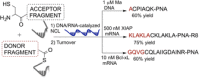
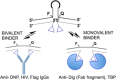
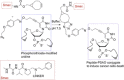
Nucleic acid directed bioorthogonal reactions offer the fascinating opportunity to unveil and redirect a plethora of intracellular mechanisms. Nano- to picomolar amounts of specific RNA molecules serve as templates and catalyze the selective formation of molecules that 1) exert biological effects, or 2) provide measurable signals for RNA detection. Turnover of reactants on the template is a valuable asset when concentrations of RNA templates are low. The idea is to use RNA-templated reactions to fully control the biodistribution of drugs and to push the detection limits of DNA or RNA analytes to extraordinary sensitivities. Herein we review recent and instructive examples of conditional synthesis or release of compounds for in cellulo protein interference and intracellular nucleic acid imaging.
Keywords: in situ drug release; in situ drug synthesis; nucleic acid encoded reactions; nucleic acid sensing.
© 2017 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Figures





Similar articles
-
Reducing product inhibition in nucleic acid-templated ligation reactions: DNA-templated cycligation.Chembiochem. 2013 Nov 25;14(17):2322-8. doi: 10.1002/cbic.201300516. Epub 2013 Oct 9. Chembiochem. 2013. PMID: 24243697
-
Amplification by nucleic acid-templated reactions.Org Biomol Chem. 2014 May 14;12(18):2821-33. doi: 10.1039/c4ob00096j. Org Biomol Chem. 2014. PMID: 24671414 Review.
-
Nucleic acid encoding to program self-assembly in chemical biology.Chem Soc Rev. 2008 Jul;37(7):1330-6. doi: 10.1039/b706610b. Epub 2008 Apr 17. Chem Soc Rev. 2008. PMID: 18568159 Review.
-
RNA-templated chemical synthesis of proapoptotic L- and d-peptides.Bioorg Med Chem. 2022 Jul 15;66:116786. doi: 10.1016/j.bmc.2022.116786. Epub 2022 May 14. Bioorg Med Chem. 2022. PMID: 35594647
-
Nucleic acid templated reactions: consequences of probe reactivity and readout strategy for amplified signaling and sequence selectivity.Chemistry. 2009 Jul 6;15(27):6723-30. doi: 10.1002/chem.200900025. Chemistry. 2009. PMID: 19496097
Cited by
-
Late-Stage Functionalization of Living Organisms: Rethinking Selectivity in Biology.Chem Rev. 2024 Feb 14;124(3):889-928. doi: 10.1021/acs.chemrev.3c00579. Epub 2024 Jan 17. Chem Rev. 2024. PMID: 38231473 Free PMC article. Review.
-
Advances in Tetrazine Bioorthogonal Chemistry Driven by the Synthesis of Novel Tetrazines and Dienophiles.Acc Chem Res. 2018 May 15;51(5):1249-1259. doi: 10.1021/acs.accounts.8b00062. Epub 2018 Apr 11. Acc Chem Res. 2018. PMID: 29638113 Free PMC article.
-
Enhancing native chemical ligation for challenging chemical protein syntheses.Curr Opin Chem Biol. 2020 Oct;58:37-44. doi: 10.1016/j.cbpa.2020.04.003. Epub 2020 Jul 31. Curr Opin Chem Biol. 2020. PMID: 32745915 Free PMC article. Review.
-
PNA-Based MicroRNA Detection Methodologies.Molecules. 2020 Mar 12;25(6):1296. doi: 10.3390/molecules25061296. Molecules. 2020. PMID: 32178411 Free PMC article. Review.
-
Information propagation through enzyme-free catalytic templating of DNA dimerization with weak product inhibition.Nat Chem. 2025 Aug;17(8):1179-1187. doi: 10.1038/s41557-025-01831-x. Epub 2025 Jun 5. Nat Chem. 2025. PMID: 40473818 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

