Soft-MS and Computational Mapping of Oleuropein
- PMID: 28481240
- PMCID: PMC5454905
- DOI: 10.3390/ijms18050992
Soft-MS and Computational Mapping of Oleuropein
Abstract
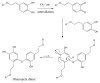
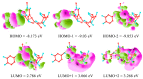
Olive oil and table olives are rich sources of biophenols, which provides a unique taste, aroma and potential health benefits. Specifically, green olive drupes are enriched with oleuropein, a bioactive biophenol secoiridoid. Olive oil contains hydrolytic derivatives such as hydroxytyrosol, oleacein and elenolate from oleuropein as well as tyrosol and oleocanthal from ligstroside. Biophenol secoiridoids are categorized by the presence of elenoic acid or its derivatives in their molecular structure. Medical studies suggest that olive biophenol secoiridoids could prevent cancer, obesity, osteoporosis, and neurodegeneration. Therefore, understanding the biomolecular dynamics of oleuropein can potentially improve olive-based functional foods and nutraceuticals. This review provides a critical assessment of oleuropein biomolecular mechanism and computational mapping that could contribute to nutrigenomics.
Keywords: biophenols; hydroxytyrosol; olive oil; secoiridoid; table olives; tyrosol; β-glucosidase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
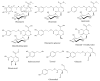
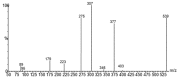
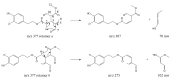
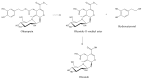


References
-
- Sivakumar G., Briccoli Bati C., Uccella N.A. HPLC-MS screening of the antioxidant profile of Italian olive cultivars. Chem. Nat. Compd. 2005;41:588–591. doi: 10.1007/s10600-005-0214-8. - DOI
-
- Sánchez de Medina V., Miho H., Melliou E., Magiatis P., Priego-Capote F., Luque de Castro M.D. Quantitative method for determination of oleocanthal and oleacein in virgin olive oils by liquid chromatography-tandem mass spectrometry. Talanta. 2017;162:24–31. doi: 10.1016/j.talanta.2016.09.056. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

