Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus
- PMID: 28481299
- PMCID: PMC5454811
- DOI: 10.3390/ijms18050887
Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus
Abstract
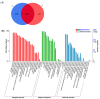
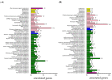
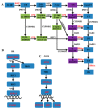
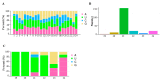
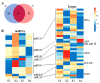
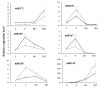
Oilseed rape (Brassica napus L.) is the second largest oilseed crop worldwide and one of the most important oil crops in China. As a component of plant architecture, branch angle plays an important role in yield performance, especially under high-density planting conditions. However, the mechanisms underlying the regulation of branch angle are still largely not understood. Two oilseed rape lines with significantly different branch angles were used to conduct RNA- and miRNA-profiling at two developmental stages, identifying differential expression of a large number of genes involved in auxin- and brassinosteroid (BR)-related pathways. Many auxin response genes, including AUX1, IAA, GH3, and ARF, were enriched in the compact line. However, a number of genes involved in BR signaling transduction and biosynthesis were down-regulated. Differentially expressed miRNAs included those involved in auxin signaling transduction. Expression patterns of most target genes were fine-tuned by related miRNAs, such as miR156, miR172, and miR319. Some miRNAs were found to be differentially expressed at both developmental stages, including three miR827 members. Our results provide insight that auxin- and BR-signaling may play a pivotal role in branch angle regulation.
Keywords: Brassica napus; auxin; branch angle; brassinosteroid; deep sequencing; miRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Mansfield B.D., Mumm R.H. Survey of plant density tolerance in US maize germplasm. Crop Sci. 2014;54:157–173. doi: 10.2135/cropsci2013.04.0252. - DOI
-
- Nik M.M., Babaeian M., Tavassoli A., Asgharzade A. Effect of plant density on yield and yield components of corn hybrids (Zea mays) Sci. Res. Essays. 2011;6:4821–4825.
-
- Duvick D.N. Genetic progress in yield of United States maize (Zea mays L.) Maydica. 2005;50:193–202.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

