Next-Generation Sequencing Reveals Novel Mutations in X-linked Intellectual Disability
- PMID: 28481730
- PMCID: PMC5586158
- DOI: 10.1089/omi.2017.0009
Next-Generation Sequencing Reveals Novel Mutations in X-linked Intellectual Disability
Abstract
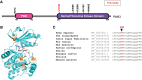
Robust diagnostics for many human genetic disorders are much needed in the pursuit of global personalized medicine. Next-generation sequencing now offers new promise for biomarker and diagnostic discovery, in developed as well as resource-limited countries. In this broader global health context, X-linked intellectual disability (XLID) is an inherited genetic disorder that is associated with a range of phenotypes impacting societies in both developed and developing countries. Although intellectual disability arises due to diverse causes, a substantial proportion is caused by genomic alterations. Studies have identified causal XLID genomic alterations in more than 100 protein-coding genes located on the X-chromosome. However, the causes for a substantial number of intellectual disability and associated phenotypes still remain unknown. Identification of causative genes and novel mutations will help in early diagnosis as well as genetic counseling of families. Advent of next-generation sequencing methods has accelerated the discovery of new genes involved in mental health disorders. In this study, we analyzed the exomes of three families from India with nonsyndromic XLID comprising seven affected individuals. The affected individuals had varying degrees of intellectual disability, microcephaly, and delayed motor and language milestones. We identified potential causal variants in three XLID genes, including PAK3 (V294M), CASK (complex structural variant), and MECP2 (P354T). Our findings reported in this study extend the spectrum of mutations and phenotypes associated with XLID, and calls for further studies of intellectual disability and mental health disorders with use of next-generation sequencing technologies.
Keywords: diagnostic medicine; genotype–phenotype association; mental retardation; neurodevelopmental disorders; next-generation sequencing.
Conflict of interest statement
Genentech authors hold Roche shares. The other authors declare no conflicts of interest.
Figures


References
-
- Arias-Romero LE, and Chernoff J. (2008). A tale of two Paks. European Cell Biol Org 100, 97–108 - PubMed
-
- Bartenhagen C, and Dugas M. (2016). Robust and exact structural variation detection with paired-end and soft-clipped alignments: SoftSV compared with eight algorithms. Brief Bioinform 17, 51–62 - PubMed
-
- Belichenko PV, Wright EE, Belichenko NP, et al. (2009). Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: Evidence for disruption of neuronal networks. J Comp Neurol 514, 240–258 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

