Soft Sweeps Are the Dominant Mode of Adaptation in the Human Genome
- PMID: 28482049
- PMCID: PMC5850737
- DOI: 10.1093/molbev/msx154
Soft Sweeps Are the Dominant Mode of Adaptation in the Human Genome
Abstract
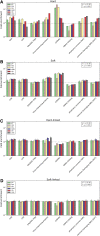
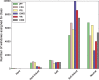
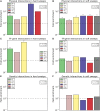
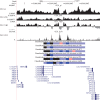
The degree to which adaptation in recent human evolution shapes genetic variation remains controversial. This is in part due to the limited evidence in humans for classic "hard selective sweeps", wherein a novel beneficial mutation rapidly sweeps through a population to fixation. However, positive selection may often proceed via "soft sweeps" acting on mutations already present within a population. Here, we examine recent positive selection across six human populations using a powerful machine learning approach that is sensitive to both hard and soft sweeps. We found evidence that soft sweeps are widespread and account for the vast majority of recent human adaptation. Surprisingly, our results also suggest that linked positive selection affects patterns of variation across much of the genome, and may increase the frequencies of deleterious mutations. Our results also reveal insights into the role of sexual selection, cancer risk, and central nervous system development in recent human evolution.
Keywords: adaptation; machine learning; population genomics; selective sweeps; soft sweeps.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Allinen M, Peri L, Kujala S, Lahti‐Domenici J, Outila K, Karppinen SM, Launonen V, Winqvist R.. 2002. Analysis of 11q21–24 loss of heterozygosity candidate target genes in breast cancer: indications of TSLC1 promoter hypermethylation. Genes Chromosomes Cancer 34:384–389. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

