Selective gene expression analysis of the neuroepithelial body microenvironment in postnatal lungs with special interest for potential stem cell characteristics
- PMID: 28482837
- PMCID: PMC5422937
- DOI: 10.1186/s12931-017-0571-4
Selective gene expression analysis of the neuroepithelial body microenvironment in postnatal lungs with special interest for potential stem cell characteristics
Abstract
Background: The pulmonary neuroepithelial body (NEB) microenvironment (ME) consists of innervated cell clusters that occur sparsely distributed in the airway epithelium, an organization that has so far hampered reliable selective gene expression analysis. Although the NEB ME has been suggested to be important for airway epithelial repair after ablation, little is known about their potential stem cell characteristics in healthy postnatal lungs. Here we report on a large-scale selective gene expression analysis of the NEB ME.
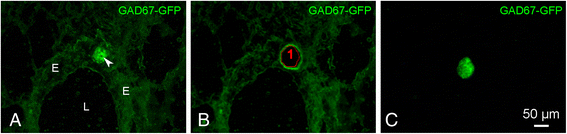
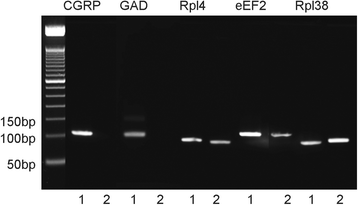
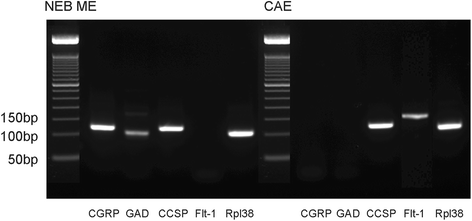
Methods: A GAD67-GFP mouse model was used that harbors GFP-fluorescent NEBs, allowing quick selection and pooling by laser microdissection (LMD) without further treatment. A panel of stem cell-related PCR arrays was used to selectively compare mRNA expression in the NEB ME to control airway epithelium (CAE). For genes that showed a higher expression in the NEB ME, a ranking was made based on the relative expression level. Single qPCR and immunohistochemistry were used to validate and quantify the PCR array data.
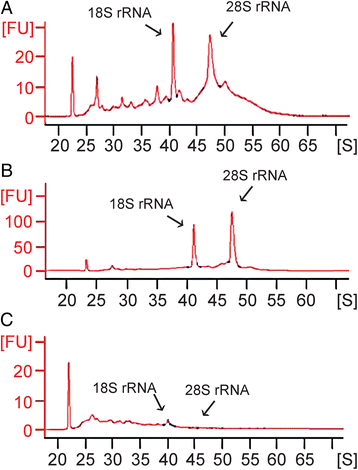
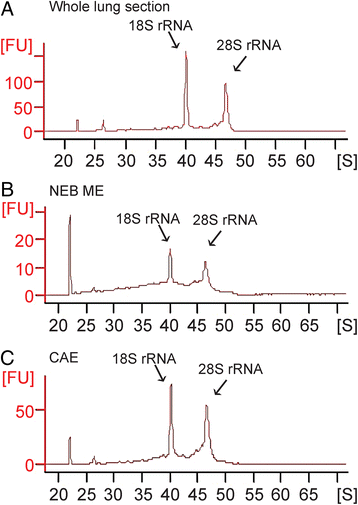
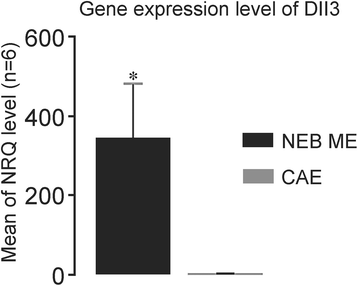
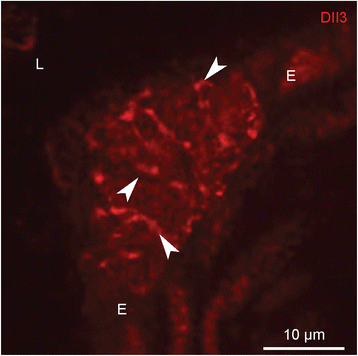
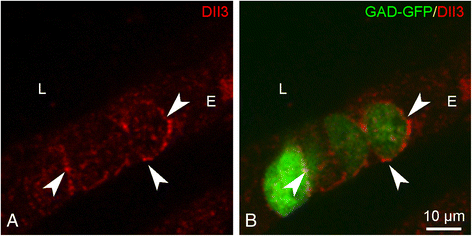
Results: Careful optimization of all protocols appeared to be essential to finally obtain high-quality RNA from pooled LMD samples of NEB ME. About 30% of the more than 600 analyzed genes showed an at least two-fold higher expression compared to CAE. The gene that showed the highest relative expression in the NEB ME, Delta-like ligand 3 (Dll3), was investigated in more detail. Selective Dll3 gene expression in the NEB ME could be quantified via single qPCR experiments, and Dll3 protein expression could be localized specifically to NEB cell surface membranes.
Conclusions: This study emphasized the importance of good protocols and RNA quality controls because of the, often neglected, fast RNA degradation in postnatal lung samples. It was shown that sufficient amounts of high-quality RNA for reliable complex gene expression analysis can be obtained from pooled LMD-collected NEB ME samples of postnatal lungs. Dll3 expression, which has also been reported to be important in high-grade pulmonary tumor-initiating cells, was used as a proof-of-concept to confirm that the described methodology represents a promising tool for further unraveling the molecular basis of NEB ME physiology in general, and its postnatal stem cell capacities in particular.
Keywords: Airway epithelium; Delta-like ligand 3; Laser microdissection; Neuroepithelial body microenvironment; PCR array; Pulmonary neuroendocrine cells; Stem cell niche.
Figures
References
-
- Stahlman MT, Gray ME. Ontogeny of neuroendocrine cells in human-fetal lung 1. An electron-microscopic study. Lab Invest. 1984;51:449–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

