A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif
- PMID: 28482907
- PMCID: PMC5422959
- DOI: 10.1186/s12977-017-0355-4
A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif
Abstract
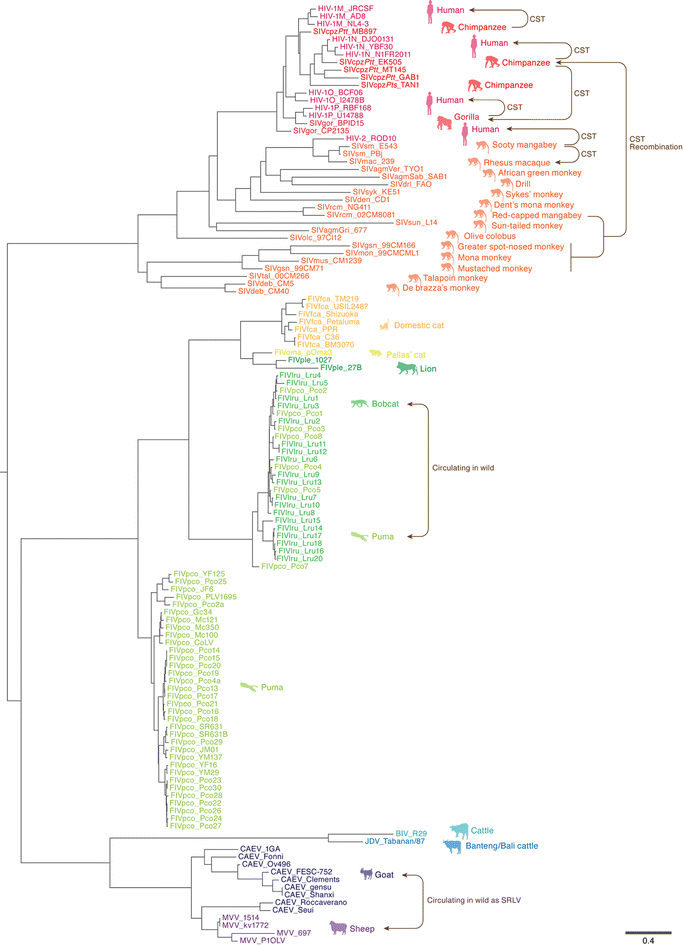
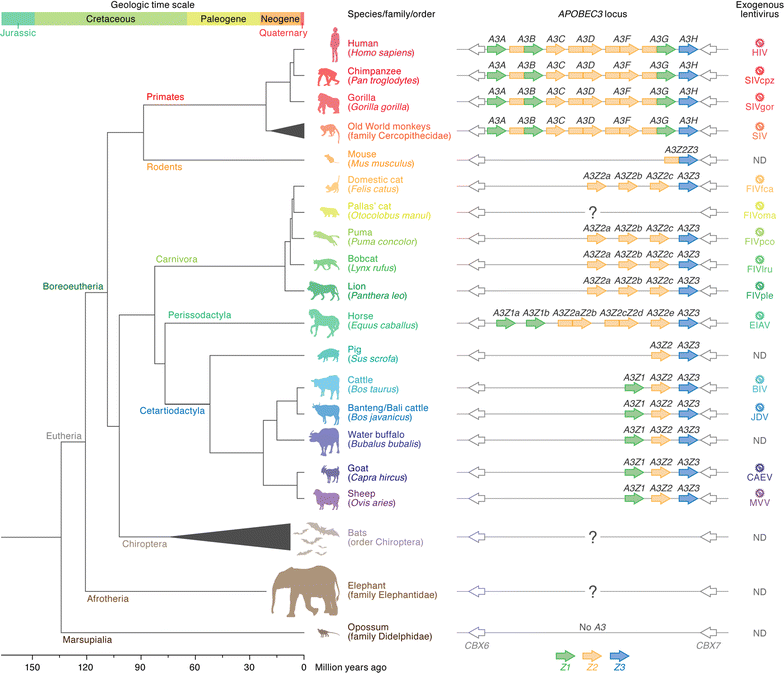
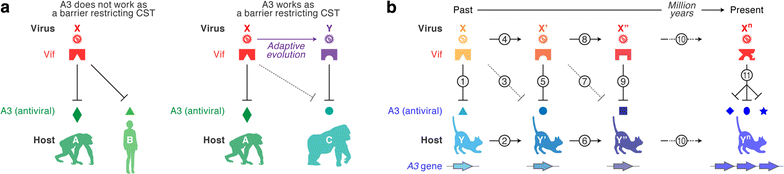
Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3) proteins are mammalian-specific cellular deaminases and have a robust ability to restrain lentivirus replication. To antagonize APOBEC3-mediated antiviral action, lentiviruses have acquired viral infectivity factor (Vif) as an accessory gene. Mammalian APOBEC3 proteins inhibit lentiviral replication by enzymatically inserting G-to-A hypermutations in the viral genome, whereas lentiviral Vif proteins degrade host APOBEC3 via the ubiquitin/proteasome-dependent pathway. Recent investigations provide evidence that lentiviral vif genes evolved to combat mammalian APOBEC3 proteins. In corollary, mammalian APOBEC3 genes are under Darwinian selective pressure to escape from antagonism by Vif. Based on these observations, it is widely accepted that lentiviral Vif and mammalian APOBEC3 have co-evolved and this concept is called an "evolutionary arms race." This review provides a comprehensive summary of current knowledge with respect to the evolutionary dynamics occurring at this pivotal host-virus interface.
Keywords: APOBEC3; Evolutionary arms race; Lentivirus; Mammal; Vif.
Figures
References
-
- Evans DT, Elder JH, Desrosiers RC. Nonhuman lentiviruses. In: Knipe DM, Howley PM, editors. Fields virology, vol. 2. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1584–1612.
-
- Freed EO, Martin MA. Human immunodeficiency viruses: replication. In: Knipe DM, Howley PM, editors. Fields virology, vol. 2. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1502–1560.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

