Processes Underlying a Reproductive Barrier in indica- japonica Rice Hybrids Revealed by Transcriptome Analysis
- PMID: 28483876
- PMCID: PMC5490891
- DOI: 10.1104/pp.17.00093
Processes Underlying a Reproductive Barrier in indica- japonica Rice Hybrids Revealed by Transcriptome Analysis
Abstract
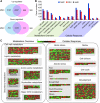
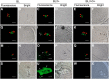
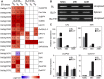
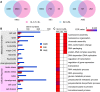
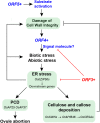
In rice (Oryza sativa), hybrids between indica and japonica subspecies are usually highly sterile, which provides a model system for studying postzygotic reproductive isolation. A killer-protector system, S5, composed of three adjacent genes (ORF3, ORF4, and ORF5), regulates female gamete fertility of indica-japonica hybrids. To characterize the processes underlying this system, we performed transcriptomic analyses of pistils from rice variety Balilla (BL), Balilla with transformed ORF5+ (BL5+) producing sterile female gametes, and Balilla with transformed ORF3+ and ORF5+ (BL3+5+) producing fertile gametes. RNA sequencing of tissues collected before (MMC), during (MEI), and after (AME) meiosis of the megaspore mother cell detected 19,269 to 20,928 genes as expressed. Comparison between BL5+ and BL showed that ORF5+ induced differential expression of 8,339, 6,278, and 530 genes at MMC, MEI, and AME, respectively. At MMC, large-scale differential expression of cell wall-modifying genes and biotic and abiotic response genes indicated that cell wall integrity damage induced severe biotic and abiotic stresses. The processes continued to MEI and induced endoplasmic reticulum (ER) stress as indicated by differential expression of ER stress-responsive genes, leading to programmed cell death at MEI and AME, resulting in abortive female gametes. In the BL3+5+/BL comparison, 3,986, 749, and 370 genes were differentially expressed at MMC, MEI, and AME, respectively. Large numbers of cell wall modification and biotic and abiotic response genes were also induced at MMC but largely suppressed at MEI without inducing ER stress and programed cell death , producing fertile gametes. These results have general implications for the understanding of biological processes underlying reproductive barriers.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

