An In Vitro TORC1 Kinase Assay That Recapitulates the Gtr-Independent Glutamine-Responsive TORC1 Activation Mechanism on Yeast Vacuoles
- PMID: 28483912
- PMCID: PMC5492174
- DOI: 10.1128/MCB.00075-17
An In Vitro TORC1 Kinase Assay That Recapitulates the Gtr-Independent Glutamine-Responsive TORC1 Activation Mechanism on Yeast Vacuoles
Abstract
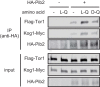
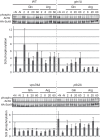
Evolutionarily conserved target of rapamycin (TOR) complex 1 (TORC1) responds to nutrients, especially amino acids, to promote cell growth. In the yeast Saccharomyces cerevisiae, various nitrogen sources activate TORC1 with different efficiencies, although the mechanism remains elusive. Leucine, and perhaps other amino acids, was reported to activate TORC1 via the heterodimeric small GTPases Gtr1-Gtr2, the orthologues of the mammalian Rag GTPases. More recently, an alternative Gtr-independent TORC1 activation mechanism that may respond to glutamine was reported, although its molecular mechanism is not clear. In studying the nutrient-responsive TORC1 activation mechanism, the lack of an in vitro assay hinders associating particular nutrient compounds with the TORC1 activation status, whereas no in vitro assay that shows nutrient responsiveness has been reported. In this study, we have developed a new in vitro TORC1 kinase assay that reproduces, for the first time, the nutrient-responsive TORC1 activation. This in vitro TORC1 assay recapitulates the previously predicted Gtr-independent glutamine-responsive TORC1 activation mechanism. Using this system, we found that this mechanism specifically responds to l-glutamine, resides on the vacuolar membranes, and involves a previously uncharacterized Vps34-Vps15 phosphatidylinositol (PI) 3-kinase complex and the PI-3-phosphate [PI(3)P]-binding FYVE domain-containing vacuolar protein Pib2. Thus, this system was proved to be useful for dissecting the glutamine-responsive TORC1 activation mechanism.
Keywords: Saccharomyces cerevisiae; TOR kinase; TORC1; Vps34; glutamine; in vitro kinase assay.
Copyright © 2017 American Society for Microbiology.
Figures

Similar articles
-
Gtr/Ego-independent TORC1 activation is achieved through a glutamine-sensitive interaction with Pib2 on the vacuolar membrane.PLoS Genet. 2018 Apr 26;14(4):e1007334. doi: 10.1371/journal.pgen.1007334. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29698392 Free PMC article.
-
A glutamine sensor that directly activates TORC1.Commun Biol. 2021 Sep 17;4(1):1093. doi: 10.1038/s42003-021-02625-w. Commun Biol. 2021. PMID: 34535752 Free PMC article.
-
Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins.J Biol Chem. 2014 Sep 5;289(36):25010-20. doi: 10.1074/jbc.M114.574335. Epub 2014 Jul 25. J Biol Chem. 2014. PMID: 25063813 Free PMC article.
-
Conserved and Divergent Mechanisms That Control TORC1 in Yeasts and Mammals.Genes (Basel). 2021 Jan 12;12(1):88. doi: 10.3390/genes12010088. Genes (Basel). 2021. PMID: 33445779 Free PMC article. Review.
-
Pib2 as an Emerging Master Regulator of Yeast TORC1.Biomolecules. 2021 Oct 9;11(10):1489. doi: 10.3390/biom11101489. Biomolecules. 2021. PMID: 34680122 Free PMC article. Review.
Cited by
-
Nutrient Signaling via the TORC1-Greatwall-PP2AB55δ Pathway Is Responsible for the High Initial Rates of Alcoholic Fermentation in Sake Yeast Strains of Saccharomyces cerevisiae.Appl Environ Microbiol. 2018 Dec 13;85(1):e02083-18. doi: 10.1128/AEM.02083-18. Print 2019 Jan 1. Appl Environ Microbiol. 2018. PMID: 30341081 Free PMC article.
-
Auxin-Inducible Depletion of the Essentialome Suggests Inhibition of TORC1 by Auxins and Inhibition of Vrg4 by SDZ 90-215, a Natural Antifungal Cyclopeptide.G3 (Bethesda). 2019 Mar 7;9(3):829-840. doi: 10.1534/g3.118.200748. G3 (Bethesda). 2019. PMID: 30670608 Free PMC article.
-
Effect of Amino Acids on Fusarium oxysporum Growth and Pathogenicity Regulated by TORC1-Tap42 Gene and Related Interaction Protein Analysis.Foods. 2023 Apr 28;12(9):1829. doi: 10.3390/foods12091829. Foods. 2023. PMID: 37174368 Free PMC article.
-
pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 ménage-à-trois to stress tolerance and longevity.Microb Cell. 2018 Jan 12;5(3):119-136. doi: 10.15698/mic2018.03.618. Microb Cell. 2018. PMID: 29487859 Free PMC article. Review.
-
Amino acid-dependent control of mTORC1 signaling: a variety of regulatory modes.J Biomed Sci. 2020 Aug 17;27(1):87. doi: 10.1186/s12929-020-00679-2. J Biomed Sci. 2020. PMID: 32799865 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
