Comparative Analysis of Extended-Spectrum-β-Lactamase CTX-M-65-Producing Salmonella enterica Serovar Infantis Isolates from Humans, Food Animals, and Retail Chickens in the United States
- PMID: 28483962
- PMCID: PMC5487606
- DOI: 10.1128/AAC.00488-17
Comparative Analysis of Extended-Spectrum-β-Lactamase CTX-M-65-Producing Salmonella enterica Serovar Infantis Isolates from Humans, Food Animals, and Retail Chickens in the United States
Abstract
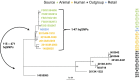
We sequenced the genomes of 10 Salmonella enterica serovar Infantis isolates containing blaCTX-M-65 obtained from chicken, cattle, and human sources collected between 2012 and 2015 in the United States through routine National Antimicrobial Resistance Monitoring System (NARMS) surveillance and product sampling programs. We also completely assembled the plasmids from four of the isolates. All isolates had a D87Y mutation in the gyrA gene and harbored between 7 and 10 resistance genes [aph(4)-Ia, aac(3)-IVa, aph(3')-Ic, blaCTX-M-65, fosA3, floR, dfrA14, sul1, tetA, aadA1] located in two distinct sites of a megaplasmid (∼316 to 323 kb) similar to that described in a blaCTX-M-65-positive S Infantis isolate from a patient in Italy. High-quality single nucleotide polymorphism (hqSNP) analysis revealed that all U.S. isolates were closely related, separated by only 1 to 38 pairwise high-quality SNPs, indicating a high likelihood that strains from humans, chickens, and cattle recently evolved from a common ancestor. The U.S. isolates were genetically similar to the blaCTX-M-65-positive S Infantis isolate from Italy, with a separation of 34 to 47 SNPs. This is the first report of the blaCTX-M-65 gene and the pESI (plasmid for emerging S Infantis)-like megaplasmid from S Infantis in the United States, and it illustrates the importance of applying a global One Health human and animal perspective to combat antimicrobial resistance.
Keywords: Salmonella; antibiotic resistance; foodborne pathogens; multidrug resistance; β-lactamases.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, Aarestrup FM. 2011. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 8:887–900. doi:10.1089/fpd.2010.0787. - DOI - PubMed
-
- Crim SM, Griffin PM, Tauxe R, Marder EP, Gilliss D, Cronquist AB, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K, Lathrop S, Zansky S, Cieslak PR, Dunn J, Holt KG, Wolpert B, Henao OL, Centers for Disease Control and Prevention. 2015. Preliminary incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2014. MMWR Morb Mortal Wkly Rep 64:495–499. - PMC - PubMed
-
- Nogrady N, Kardos G, Bistyak A, Turcsanyi I, Meszaros J, Galantai Z, Juhasz A, Samu P, Kaszanyitzky JE, Paszti J, Kiss I. 2008. Prevalence and characterization of Salmonella infantis isolates originating from different points of the broiler chicken-human food chain in Hungary. Int J Food Microbiol 127:162–167. doi:10.1016/j.ijfoodmicro.2008.07.005. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

