Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors
- PMID: 28484005
- PMCID: PMC5474804
- DOI: 10.1073/pnas.1617743114
Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors
Abstract
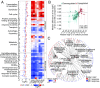
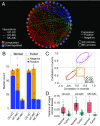
Tumors of distinct tissues of origin and genetic makeup display common hallmark cellular phenotypes, including sustained proliferation, suppression of cell death, and altered metabolism. These phenotypic commonalities have been proposed to stem from disruption of conserved regulatory mechanisms evolved during the transition to multicellularity to control fundamental cellular processes such as growth and replication. Dating the evolutionary emergence of human genes through phylostratigraphy uncovered close association between gene age and expression level in RNA sequencing data from The Cancer Genome Atlas for seven solid cancers. Genes conserved with unicellular organisms were strongly up-regulated, whereas genes of metazoan origin were primarily inactivated. These patterns were most consistent for processes known to be important in cancer, implicating both selection and active regulation during malignant transformation. The coordinated expression of strongly interacting multicellularity and unicellularity processes was lost in tumors. This separation of unicellular and multicellular functions appeared to be mediated by 12 highly connected genes, marking them as important general drivers of tumorigenesis. Our findings suggest common principles closely tied to the evolutionary history of genes underlie convergent changes at the cellular process level across a range of solid cancers. We propose altered activity of genes at the interfaces between multicellular and unicellular regions of human gene regulatory networks activate primitive transcriptional programs, driving common hallmark features of cancer. Manipulation of cross-talk between biological processes of different evolutionary origins may thus present powerful and broadly applicable treatment strategies for cancer.
Keywords: atavism; cancer; evolution; systems biology; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Ancestral gene regulatory networks drive cancer.Proc Natl Acad Sci U S A. 2017 Jun 13;114(24):6160-6162. doi: 10.1073/pnas.1706990114. Epub 2017 Jun 5. Proc Natl Acad Sci U S A. 2017. PMID: 28584134 Free PMC article. No abstract available.
Similar articles
-
How the evolution of multicellularity set the stage for cancer.Br J Cancer. 2018 Jan;118(2):145-152. doi: 10.1038/bjc.2017.398. Epub 2018 Jan 16. Br J Cancer. 2018. PMID: 29337961 Free PMC article. Review.
-
Somatic mutations in early metazoan genes disrupt regulatory links between unicellular and multicellular genes in cancer.Elife. 2019 Feb 26;8:e40947. doi: 10.7554/eLife.40947. Elife. 2019. PMID: 30803482 Free PMC article.
-
Disruption of metazoan gene regulatory networks in cancer alters the balance of co-expression between genes of unicellular and multicellular origins.Genome Biol. 2024 Apr 29;25(1):110. doi: 10.1186/s13059-024-03247-1. Genome Biol. 2024. PMID: 38685127 Free PMC article.
-
Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data.Semin Cancer Biol. 2015 Feb;30:4-12. doi: 10.1016/j.semcancer.2014.04.002. Epub 2014 Apr 18. Semin Cancer Biol. 2015. PMID: 24747696 Review.
-
Reverting to single-cell biology: The predictions of the atavism theory of cancer.Prog Biophys Mol Biol. 2021 Oct;165:49-55. doi: 10.1016/j.pbiomolbio.2021.08.002. Epub 2021 Aug 8. Prog Biophys Mol Biol. 2021. PMID: 34371024 Free PMC article.
Cited by
-
Is Cancer Metabolism an Atavism?Cancers (Basel). 2024 Jun 29;16(13):2415. doi: 10.3390/cancers16132415. Cancers (Basel). 2024. PMID: 39001477 Free PMC article. Review.
-
A Thermodynamic Model for Water Activity and Redox Potential in Evolution and Development.J Mol Evol. 2022 Apr;90(2):182-199. doi: 10.1007/s00239-022-10051-7. Epub 2022 Mar 13. J Mol Evol. 2022. PMID: 35279735
-
Resolution of Complex Issues in Genome Regulation and Cancer Requires Non-Linear and Network-Based Thermodynamics.Int J Mol Sci. 2019 Dec 29;21(1):240. doi: 10.3390/ijms21010240. Int J Mol Sci. 2019. PMID: 31905791 Free PMC article. Review.
-
BID and the α-bisabolol-triggered cell death program: converging on mitochondria and lysosomes.Cell Death Dis. 2019 Nov 26;10(12):889. doi: 10.1038/s41419-019-2126-8. Cell Death Dis. 2019. PMID: 31767857 Free PMC article.
-
How the evolution of multicellularity set the stage for cancer.Br J Cancer. 2018 Jan;118(2):145-152. doi: 10.1038/bjc.2017.398. Epub 2018 Jan 16. Br J Cancer. 2018. PMID: 29337961 Free PMC article. Review.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. - PubMed
-
- Vincent M. Cancer: A de-repression of a default survival program common to all cells?: A life-history perspective on the nature of cancer. BioEssays. 2012;34:72–82. - PubMed
-
- Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

