Heritability analysis with repeat measurements and its application to resting-state functional connectivity
- PMID: 28484032
- PMCID: PMC5448225
- DOI: 10.1073/pnas.1700765114
Heritability analysis with repeat measurements and its application to resting-state functional connectivity
Abstract
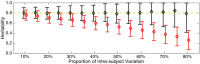
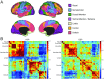
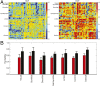
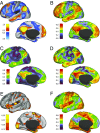
Heritability, defined as the proportion of phenotypic variation attributable to genetic variation, provides important information about the genetic basis of a trait. Existing heritability analysis methods do not discriminate between stable effects (e.g., due to the subject's unique environment) and transient effects, such as measurement error. This can lead to misleading assessments, particularly when comparing the heritability of traits that exhibit different levels of reliability. Here, we present a linear mixed effects model to conduct heritability analyses that explicitly accounts for intrasubject fluctuations (e.g., due to measurement noise or biological transients) using repeat measurements. We apply the proposed strategy to the analysis of resting-state fMRI measurements-a prototypic data modality that exhibits variable levels of test-retest reliability across space. Our results reveal that the stable components of functional connectivity within and across well-established large-scale brain networks can be considerably heritable. Furthermore, we demonstrate that dissociating intra- and intersubject variation can reveal genetic influence on a phenotype that is not fully captured by conventional heritability analyses.
Keywords: functional connectivity; heritability; repeat measurements; resting-state fMRI; test–retest reliability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Reply to Risk and Zhu: Mixed-effects modeling as a principled approach to heritability analysis with repeat measurements.Proc Natl Acad Sci U S A. 2018 Jan 9;115(2):E123. doi: 10.1073/pnas.1719882115. Epub 2017 Dec 29. Proc Natl Acad Sci U S A. 2018. PMID: 29288224 Free PMC article. No abstract available.
-
Note on bias from averaging repeated measurements in heritability studies.Proc Natl Acad Sci U S A. 2018 Jan 9;115(2):E122. doi: 10.1073/pnas.1719250115. Epub 2017 Dec 29. Proc Natl Acad Sci U S A. 2018. PMID: 29288225 Free PMC article. No abstract available.
References
-
- Visscher P, Hill W, Wray N. Heritability in the genomics era—Concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. - PubMed
-
- Falconer D. Introduction to Quantitative Genetics. Oliver and Boyd; Edinburgh: 1960.
-
- Polderman T, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–709. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 NS070963/NS/NINDS NIH HHS/United States
- K25 EB013649/EB/NIBIB NIH HHS/United States
- K24 MH094614/MH/NIMH NIH HHS/United States
- R41 AG052246/AG/NIA NIH HHS/United States
- U54 MH091657/MH/NIMH NIH HHS/United States
- K01 MH099232/MH/NIMH NIH HHS/United States
- R21 AG050122/AG/NIA NIH HHS/United States
- S10 RR023043/RR/NCRR NIH HHS/United States
- P41 EB015896/EB/NIBIB NIH HHS/United States
- R01 NS083534/NS/NINDS NIH HHS/United States
- S10 RR023401/RR/NCRR NIH HHS/United States
- R01 MH101486/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

