Evolution of sex-dependent mtDNA transmission in freshwater mussels (Bivalvia: Unionida)
- PMID: 28484275
- PMCID: PMC5431520
- DOI: 10.1038/s41598-017-01708-1
Evolution of sex-dependent mtDNA transmission in freshwater mussels (Bivalvia: Unionida)
Abstract
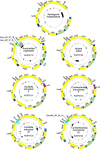
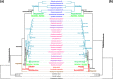
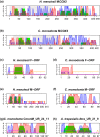
Doubly uniparental inheritance (DUI) describes a mode of mtDNA transmission widespread in gonochoric freshwater mussels (Bivalvia: Palaeoheterodonta: Unionida). In this system, both female- and male-transmitted mtDNAs, named F and M respectively, coexist in the same species. In unionids, DUI is strictly correlated to gonochorism and to the presence of the atypical open reading frames (ORFans) F-orf and M-orf, respectively inside F and M mtDNAs, which are hypothesized to participate in sex determination. However, DUI is not found in all three Unionida superfamilies (confirmed in Hyrioidea and Unionoidea but not in Etherioidea), raising the question of its origin in these bivalves. To reconstruct the co-evolution of DUI and of ORFans, we sequenced the mtDNAs of four unionids (two gonochoric with DUI, one gonochoric and one hermaphroditic without DUI) and of the related gonochoric species Neotrigonia margaritacea (Palaeoheterodonta: Trigoniida). Our analyses suggest that rearranged mtDNAs appeared early during unionid radiation, and that a duplicated and diverged atp8 gene evolved into the M-orf associated with the paternal transmission route in Hyrioidea and Unionoidea, but not in Etherioidea. We propose that novel mtDNA-encoded genes can deeply influence bivalve sex determining systems and the evolution of the mitogenomes in which they occur.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Zouros E. Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol. Biol. 2013;40:1–31. doi: 10.1007/s11692-012-9195-2. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

