GAD1 alternative transcripts and DNA methylation in human prefrontal cortex and hippocampus in brain development, schizophrenia
- PMID: 28485403
- PMCID: PMC7564279
- DOI: 10.1038/mp.2017.105
GAD1 alternative transcripts and DNA methylation in human prefrontal cortex and hippocampus in brain development, schizophrenia
Abstract
Genetic variations and adverse environmental events in utero or shortly after birth can lead to abnormal brain development and increased risk of schizophrenia. γ-Aminobutyric acid (GABA), the major inhibitory neurotransmitter in the mammalian brain, plays a vital role in normal brain development. GABA synthesis is controlled by enzymes derived from two glutamic acid decarboxylase (GAD) genes, GAD1 and GAD2, both of which produce transcript isoforms. While the full-length GAD1 transcript (GAD67) has been implicated in the neuropathology of schizophrenia, the transcript structure of GAD1 in the human brain has not been fully characterized. In this study, with the use of RNA sequencing and PCR technologies, we report the discovery of 10 novel transcripts of GAD1 in the human brain. Expression levels of four novel GAD1 transcripts (8A, 8B, I80 and I86) showed a lifespan trajectory expression pattern that is anticorrelated with the expression of the full-length GAD1 transcript. In addition, methylation levels of two CpG loci within the putative GAD1 promoter were significantly associated with the schizophrenia-risk SNP rs3749034 and with the expression of GAD25 in dorsolateral prefrontal cortex (DLPFC). Moreover, schizophrenia patients who had completed suicide and/or were positive for nicotine exposure had significantly higher full-length GAD1 expression in the DLPFC. Alternative splicing of GAD1 and epigenetic state appear to play roles in the developmental profile of GAD1 expression and may contribute to GABA dysfunction in the PFC and hippocampus of patients with schizophrenia.
Conflict of interest statement
Conflict of Interest
The authors of this paper have no conflicts of interest to disclose.
Figures

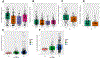
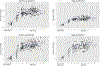
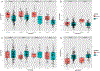
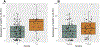
References
-
- LoTurco JJ, Owens DF, Heath MJ, Davis MB, Kriegstein AR. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron 1995; 15: 1287–1298. - PubMed
-
- Marty S, Berninger B, Carroll P, Thoenen H. GABAergic stimulation regulates the phenotype of hippocampal interneurons through the regulation of brain-derived neurotrophic factor. Neuron 1996; 16: 565–570. - PubMed
-
- Behar TN, Schaffner AE, Scott CA, Greene CL, Barker JL. GABA receptor antagonists modulate postmitotic cell migration in slice cultures of embryonic rat cortex. Cereb Cortex N Y N 1991 2000; 10: 899–909. - PubMed
-
- Varju P, Katarova Z, Madarász E, Szabó G. Sequential induction of embryonic and adult forms of glutamic acid decarboxylase during in vitro-induced neurogenesis in cloned neuroectodermal cell-line, NE-7C2. J Neurochem 2002; 80: 605–615. - PubMed
-
- Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron 1991; 7: 91–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

