Structure-Activity Relationship of 2,4-Dichloro-N-(3,5-dichloro-4-(quinolin-3-yloxy)phenyl)benzenesulfonamide (INT131) Analogs for PPARγ-Targeted Antidiabetics
- PMID: 28485590
- PMCID: PMC5537074
- DOI: 10.1021/acs.jmedchem.6b01727
Structure-Activity Relationship of 2,4-Dichloro-N-(3,5-dichloro-4-(quinolin-3-yloxy)phenyl)benzenesulfonamide (INT131) Analogs for PPARγ-Targeted Antidiabetics
Abstract
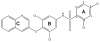
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor central to fatty acid and glucose homeostasis. PPARγ is the molecular target for type 2 diabetes mellitus (T2DM) therapeutics TZDs (thiazolidinediones), full agonists of PPARγ with robust antidiabetic properties, which are confounded with significant side effects. Partial agonists of PPARγ, such as INT131 (1), have displayed similar insulin-sensitizing efficacy as TZDs, but lack many side effects. To probe the structure-activity relationship (SAR) of the scaffold 1, we synthesized 14 analogs of compound 1 which revealed compounds with higher transcriptional potency for PPARγ and identification of moieties of the scaffold 1 key to high transcriptional potency. The sulfonamide linker is critical to activity, substitutions at position 4 of the benzene ring A were associated with higher transcriptional activity, substitutions at position 2 aided in tighter packing and activity, and the ring type and size of ring A affected the degree of activity.
Conflict of interest statement
Figures





References
-
- Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y, Nettles KW, Griffin PR. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. - PubMed
-
- Taygerly JP, McGee LR, Rubenstein SM, Houze JB, Cushing TD, Li Y, Motani A, Chen JL, Frankmoelle W, Ye G, Learned MR, Jaen J, Miao S, Timmermans PB, Thoolen M, Kearney P, Flygare J, Beckmann H, Weiszmann J, Lindstrom M, Walker N, Liu J, Biermann D, Wang Z, Hagiwara A, Iida T, Aramaki H, Kitao Y, Shinkai H, Furukawa N, Nishiu J, Nakamura M. Discovery of INT131: a selective PPARgamma modulator that enhances insulin sensitivity. Bioorg Med Chem. 2013;21:979–992. - PubMed
-
- Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O'Rahilly S. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

