Super-resolution imaging of a 2.5 kb non-repetitive DNA in situ in the nuclear genome using molecular beacon probes
- PMID: 28485713
- PMCID: PMC5433842
- DOI: 10.7554/eLife.21660
Super-resolution imaging of a 2.5 kb non-repetitive DNA in situ in the nuclear genome using molecular beacon probes
Abstract
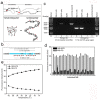
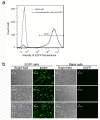
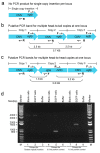
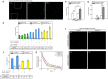
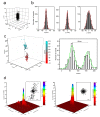
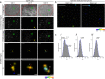
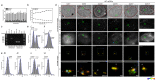
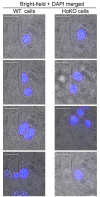
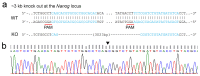
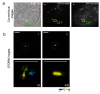
High-resolution visualization of short non-repetitive DNA in situ in the nuclear genome is essential for studying looping interactions and chromatin organization in single cells. Recent advances in fluorescence in situ hybridization (FISH) using Oligopaint probes have enabled super-resolution imaging of genomic domains with a resolution limit of 4.9 kb. To target shorter elements, we developed a simple FISH method that uses molecular beacon (MB) probes to facilitate the probe-target binding, while minimizing non-specific fluorescence. We used three-dimensional stochastic optical reconstruction microscopy (3D-STORM) with optimized imaging conditions to efficiently distinguish sparsely distributed Alexa-647 from background cellular autofluorescence. Utilizing 3D-STORM and only 29-34 individual MB probes, we observed 3D fine-scale nanostructures of 2.5 kb integrated or endogenous unique DNA in situ in human or mouse genome, respectively. We demonstrated our MB-based FISH method was capable of visualizing the so far shortest non-repetitive genomic sequence in 3D at super-resolution.
Keywords: chromatin organization; chromosomes; fluorescence in situ hybridization; genes; molecular beacons; non-repetitive DNA; none; stochastic optical reconstruction microscopy; super-resolution imaging.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


References
-
- Ahuja AK, Jodkowska K, Teloni F, Bizard AH, Zellweger R, Herrador R, Ortega S, Hickson ID, Altmeyer M, Mendez J, Lopes M. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nature Communications. 2016;7:10660. doi: 10.1038/ncomms10660. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

