Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models
- PMID: 28485718
- PMCID: PMC5423390
- DOI: 10.1038/sdata.2017.61
Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models
Abstract
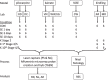
Global expression profiling of neurologic or psychiatric disorders has been confounded by variability among laboratories, animal models, tissues sampled, and experimental platforms, with the result being that few genes demonstrate consistent expression changes. We attempted to minimize these confounds by pooling dentate granule cell transcriptional profiles from 164 rats in seven laboratories, using three status epilepticus (SE) epilepsy models (pilocarpine, kainate, self-sustained SE), plus amygdala kindling. In each epilepsy model, RNA was harvested from laser-captured dentate granule cells from six rats at four time points early in the process of developing epilepsy, and data were collected from two independent laboratories in each rodent model except SSSE. Hierarchical clustering of differentially-expressed transcripts in the three SE models revealed complete separation between controls and SE rats isolated 1 day after SE. However, concordance of gene expression changes in the SE models was only 26-38% between laboratories, and 4.5% among models, validating the consortium approach. Transcripts with unusually highly variable control expression across laboratories provide a 'red herring' list for low-powered studies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Comment in
-
A Look Behind the Curtain: Epilepsy Microarray Consortium.Epilepsy Curr. 2017 Nov-Dec;17(6):374-376. doi: 10.5698/1535-7597.17.6.374. Epilepsy Curr. 2017. PMID: 29217985 Free PMC article. No abstract available.
References
Data Citations
-
- Dingledine R. 2013. NCBI Gene Expression Omnibus. GSE47752
References
-
- Hesdorffer D. C., Logroscino G., Cascino G., Annegers J. F. & Hauser W. A. Risk of unprovoked seizure after acute symptomatic seizure: effect of status epilepticus. Annals of Neurology 44, 908–912 (1998). - PubMed
-
- Lowenstein D. H., Bleck T. & Macdonald R. L. It's time to revise the definition of status epilepticus. Epilepsia 40, 120–122 (1999). - PubMed
-
- Raspall-Chaure M., Chin R. F., Neville B. G. & Scott R. C. Outcome of paediatric convulsive status epilepticus: a systematic review. The Lancet Neurology 5, 769–779 (2006). - PubMed
-
- Lukasiuk K., Kontula L. & Pitkänen A. cDNA profiling of epileptogenesis in the rat brain. Eur J Neurosci. 17, 271–279 (2003). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

