A new model for ancient DNA decay based on paleogenomic meta-analysis
- PMID: 28486705
- PMCID: PMC5499742
- DOI: 10.1093/nar/gkx361
A new model for ancient DNA decay based on paleogenomic meta-analysis
Abstract
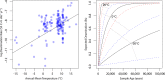
The persistence of DNA over archaeological and paleontological timescales in diverse environments has led to a revolutionary body of paleogenomic research, yet the dynamics of DNA degradation are still poorly understood. We analyzed 185 paleogenomic datasets and compared DNA survival with environmental variables and sample ages. We find cytosine deamination follows a conventional thermal age model, but we find no correlation between DNA fragmentation and sample age over the timespans analyzed, even when controlling for environmental variables. We propose a model for ancient DNA decay wherein fragmentation rapidly reaches a threshold, then subsequently slows. The observed loss of DNA over time may be due to a bulk diffusion process in many cases, highlighting the importance of tissues and environments creating effectively closed systems for DNA preservation. This model of DNA degradation is largely based on mammal bone samples due to published genomic dataset availability. Continued refinement to the model to reflect diverse biological systems and tissue types will further improve our understanding of ancient DNA breakdown dynamics.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Shapiro B., Hofreiter M.. A paleogenomic perspective on evolution and gene function: new insights from ancient DNA. Science. 2014; 343:1236573–1236573. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

