A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy
- PMID: 28486927
- PMCID: PMC5424349
- DOI: 10.1186/s12859-017-1670-4
A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy
Abstract
Background: Species-level classification for 16S rRNA gene sequences remains a serious challenge for microbiome researchers, because existing taxonomic classification tools for 16S rRNA gene sequences either do not provide species-level classification, or their classification results are unreliable. The unreliable results are due to the limitations in the existing methods which either lack solid probabilistic-based criteria to evaluate the confidence of their taxonomic assignments, or use nucleotide k-mer frequency as the proxy for sequence similarity measurement.
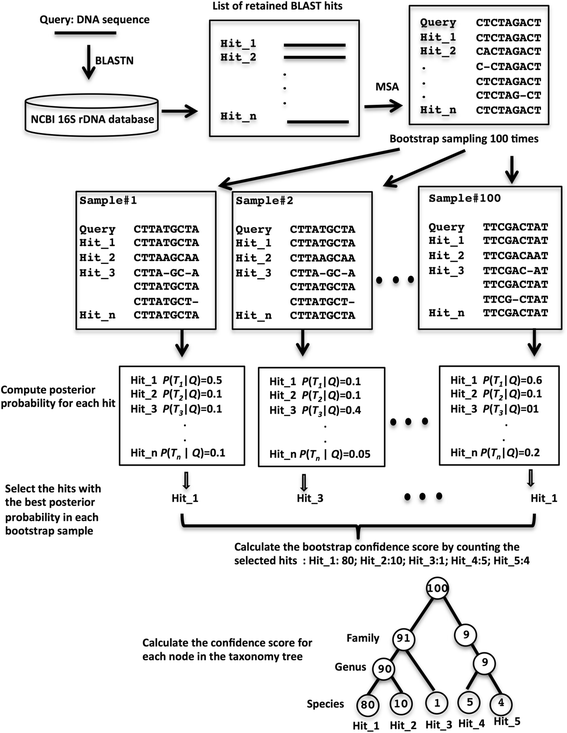
Results: We have developed a method that shows significantly improved species-level classification results over existing methods. Our method calculates true sequence similarity between query sequences and database hits using pairwise sequence alignment. Taxonomic classifications are assigned from the species to the phylum levels based on the lowest common ancestors of multiple database hits for each query sequence, and further classification reliabilities are evaluated by bootstrap confidence scores. The novelty of our method is that the contribution of each database hit to the taxonomic assignment of the query sequence is weighted by a Bayesian posterior probability based upon the degree of sequence similarity of the database hit to the query sequence. Our method does not need any training datasets specific for different taxonomic groups. Instead only a reference database is required for aligning to the query sequences, making our method easily applicable for different regions of the 16S rRNA gene or other phylogenetic marker genes.
Conclusions: Reliable species-level classification for 16S rRNA or other phylogenetic marker genes is critical for microbiome research. Our software shows significantly higher classification accuracy than the existing tools and we provide probabilistic-based confidence scores to evaluate the reliability of our taxonomic classification assignments based on multiple database matches to query sequences. Despite its higher computational costs, our method is still suitable for analyzing large-scale microbiome datasets for practical purposes. Furthermore, our method can be applied for taxonomic classification of any phylogenetic marker gene sequences. Our software, called BLCA, is freely available at https://github.com/qunfengdong/BLCA .
Keywords: 16S rRNA gene; Taxonomic classification.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous