Structure-function analysis of RBP-J-interacting and tubulin-associated (RITA) reveals regions critical for repression of Notch target genes
- PMID: 28487372
- PMCID: PMC5481562
- DOI: 10.1074/jbc.M117.791707
Structure-function analysis of RBP-J-interacting and tubulin-associated (RITA) reveals regions critical for repression of Notch target genes
Abstract
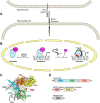
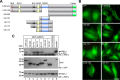
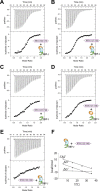
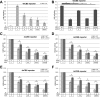
The Notch pathway is a cell-to-cell signaling mechanism that is essential for tissue development and maintenance, and aberrant Notch signaling has been implicated in various cancers, congenital defects, and cardiovascular diseases. Notch signaling activates the expression of target genes, which are regulated by the transcription factor CSL (CBF1/RBP-J, Su(H), Lag-1). CSL interacts with both transcriptional corepressor and coactivator proteins, functioning as both a repressor and activator, respectively. Although Notch activation complexes are relatively well understood at the structural level, less is known about how CSL interacts with corepressors. Recently, a new RBP-J (mammalian CSL ortholog)-interacting protein termed RITA has been identified and shown to export RBP-J out of the nucleus, thereby leading to the down-regulation of Notch target gene expression. However, the molecular details of RBP-J/RITA interactions are unclear. Here, using a combination of biochemical/cellular, structural, and biophysical techniques, we demonstrate that endogenous RBP-J and RITA proteins interact in cells, map the binding regions necessary for RBP-J·RITA complex formation, and determine the X-ray structure of the RBP-J·RITA complex bound to DNA. To validate the structure and glean more insights into function, we tested structure-based RBP-J and RITA mutants with biochemical/cellular assays and isothermal titration calorimetry. Whereas our structural and biophysical studies demonstrate that RITA binds RBP-J similarly to the RAM (RBP-J-associated molecule) domain of Notch, our biochemical and cellular assays suggest that RITA interacts with additional regions in RBP-J. Taken together, these results provide molecular insights into the mechanism of RITA-mediated regulation of Notch signaling, contributing to our understanding of how CSL functions as a transcriptional repressor of Notch target genes.
Keywords: Notch pathway; X-ray crystallography; gene regulation; isothermal titration calorimetry (ITC); protein-protein interaction; signal transduction; transcription corepressor; transcription factor.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Bray S. J. (2016) Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735 - PubMed
-
- Swiatek P. J., Lindsell C. E., del Amo F. F., Weinmaster G., and Gridley T. (1994) Notch1 is essential for postimplantation development in mice. Genes Dev. 8, 707–719 - PubMed
-
- Gridley T. (2003) Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 12, R9–R13 - PubMed
-
- Koch U., and Radtke F. (2010) Notch signaling in solid tumors. Curr. Top. Dev. Biol. 92, 411–455 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

