Functional genomics in Brugia malayi reveal diverse muscle nAChRs and differences between cholinergic anthelmintics
- PMID: 28487481
- PMCID: PMC5448196
- DOI: 10.1073/pnas.1619820114
Functional genomics in Brugia malayi reveal diverse muscle nAChRs and differences between cholinergic anthelmintics
Abstract
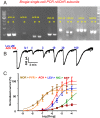
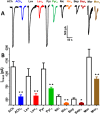
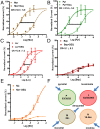
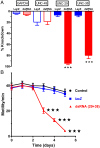
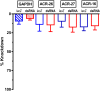
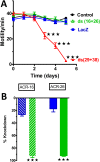
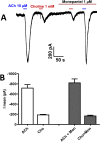
Many techniques for studying functional genomics of important target sites of anthelmintics have been restricted to Caenorhabditis elegans because they have failed when applied to animal parasites. To overcome these limitations, we have focused our research on the human nematode parasite Brugia malayi, which causes elephantiasis. Here, we combine single-cell PCR, whole muscle cell patch clamp, motility phenotyping (Worminator), and dsRNA for RNAi for functional genomic studies that have revealed, in vivo, four different muscle nAChRs (M-, L-, P-, and N-). The cholinergic anthelmintics had different selectivities for these receptors. We show that motility and patch-clamp responses to levamisole and pyrantel, but not morantel or nicotine, require the unc-38 and/or unc-29 genes. Derquantel behaved as a competitive antagonist and distinguished M-nAChRs activated by morantel (Kb 13.9 nM), P-nAChRs activated by pyrantel (Kb 126 nM), and L-nAChRs activated by levamisole (Kb 0.96 µM) and bephenium. Derquantel was a noncompetitive antagonist of nicotine, revealing N-type nAChRs. The presence of four diverse nAChRs on muscle is perhaps surprising and not predicted from the C. elegans model. The diverse nAChRs represent distinguishable drug targets with different functions: Knockdown of unc-38+unc-29 (L- and/or P-receptors) inhibited motility but knockdown of acr-16+acr-26 (M- and/or N-receptors) did not.
Keywords: Brugia; dsRNA; filaria; nAChR; patch clamp.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

