Effects of Inhibitors on the Transcriptional Profiling of Gluconobater oxydans NL71 Genes after Biooxidation of Xylose into Xylonate
- PMID: 28487685
- PMCID: PMC5403930
- DOI: 10.3389/fmicb.2017.00716
Effects of Inhibitors on the Transcriptional Profiling of Gluconobater oxydans NL71 Genes after Biooxidation of Xylose into Xylonate
Abstract
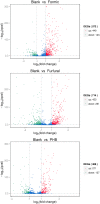
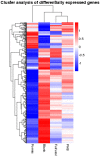
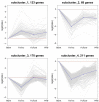
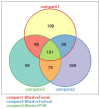
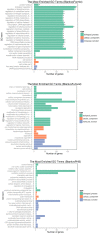
D-Xylonic acid belongs to the top 30 biomass-based platform chemicals and represents a promising application of xylose. Until today, Gluconobacter oxydans NL71 is the most efficient microbe capable of fermenting xylose into xylonate. However, its growth is seriously inhibited when concentrated lignocellulosic hydrolysates are used as substrates due to the presence of various degraded compounds formed during biomass pretreatment. Three critical lignocellulosic inhibitors were thereby identified, i.e., formic acid, furfural, and 4-hydroxybenzaldehyde. As microbe fermentation is mostly regulated at the genome level, four groups of cell transcriptomes were obtained for a comparative investigation by RNA sequencing of a control sample with samples treated separately with the above-mentioned inhibitors. The digital gene expression profiles screened 572, 714 genes, and 408 DEGs was obtained by the comparisons among four transcriptomes. A number of genes related to the different functional groups showed characteristic expression patterns induced by three inhibitors, in which 19 genes were further tested and confirmed by qRT-PCR. We extrapolated many differentially expressed genes that could explain the cellular responses to the inhibitory effects. We provide results that enable the scientific community to better define the molecular processes involved in the microbes' responses to lignocellulosic inhibitors during the cellular biooxidation of xylose into xylonic acid.
Keywords: Gluconobacter oxydans; lignocellulosic inhibitor; responsible gene; transcriptome sequencing; xylonate; xylose.
Figures
References
-
- Adachi O., Ano Y., Toyama H., Matsushita K. (2007). Chapter 1: Biooxidation with PQQ- and FAD-dependent dehydrogenases, in Modern Biooxidation: Enzymes, Reactions and Applications, eds Schmid R. D., Urlacher V. B. (Weinheim: Wiley-VCH; ), 1–41.
-
- Asai T., Iizuka H., Komagata K. (1964). The flagellation and taxonomy of genera Gluconobacter and Acetobacter with reference to the existence of intermediate strains. J. Gen. Appl. Microbiol. 10, 95–125. 10.2323/jgam.10.95 - DOI
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol). 57, 289–300.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

