Evolutionary Biology Needs Wild Microbiomes
- PMID: 28487687
- PMCID: PMC5404107
- DOI: 10.3389/fmicb.2017.00725
Evolutionary Biology Needs Wild Microbiomes
Abstract
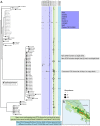
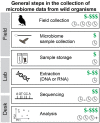
The microbiome is a vital component to the evolution of a host and much of what we know about the microbiome derives from studies on humans and captive animals. But captivity alters the microbiome and mammals have unique biological adaptations that affect their microbiomes (e.g., milk). Birds represent over 30% of known tetrapod diversity and possess their own suite of adaptations relevant to the microbiome. In a previous study, we showed that 59 species of birds displayed immense variation in their microbiomes and host (bird) taxonomy and ecology were most correlated with the gut microbiome. In this Frontiers Focused Review, I put those results in a broader context by discussing how collecting and analyzing wild microbiomes contributes to the main goals of evolutionary biology and the specific ways that birds are unique microbial hosts. Finally, I outline some of the methodological considerations for adding microbiome sampling to the research of wild animals and urge researchers to do so. To truly understand the evolution of a host, we need to understand the millions of microorganisms that inhabit it as well: evolutionary biology needs wild microbiomes.
Keywords: evolution; field biology; gut microbiome; host-associated microbiota; ornithology.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

