Gamma-Glutamylpolyamine Synthetase GlnA3 Is Involved in the First Step of Polyamine Degradation Pathway in Streptomyces coelicolor M145
- PMID: 28487688
- PMCID: PMC5403932
- DOI: 10.3389/fmicb.2017.00726
Gamma-Glutamylpolyamine Synthetase GlnA3 Is Involved in the First Step of Polyamine Degradation Pathway in Streptomyces coelicolor M145
Abstract
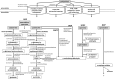
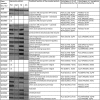
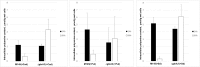
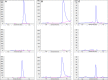
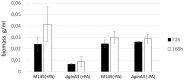
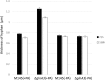
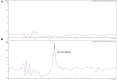
Streptomyces coelicolor M145 was shown to be able to grow in the presence of high concentrations of polyamines, such as putrescine, cadaverine, spermidine, or spermine, as a sole nitrogen source. However, hardly anything is known about polyamine utilization and its regulation in streptomycetes. In this study, we demonstrated that only one of the three proteins annotated as glutamine synthetase-like protein, GlnA3 (SCO6962), was involved in the catabolism of polyamines. Transcriptional analysis revealed that the expression of glnA3 was strongly induced by exogenous polyamines and repressed in the presence of ammonium. The ΔglnA3 mutant was shown to be unable to grow on defined Evans agar supplemented with putrescine, cadaverine, spermidine, and spermine as sole nitrogen source. HPLC analysis demonstrated that the ΔglnA3 mutant accumulated polyamines intracellularly, but was unable to degrade them. In a rich complex medium supplemented with a mixture of the four different polyamines, the ΔglnA3 mutant grew poorly showing abnormal mycelium morphology and decreased life span in comparison to the parental strain. These observations indicated that the accumulation of polyamines was toxic for the cell. An in silico analysis of the GlnA3 protein model suggested that it might act as a gamma-glutamylpolyamine synthetase catalyzing the first step of polyamine degradation. GlnA3-catalyzed glutamylation of putrescine was confirmed in an enzymatic in vitro assay and the GlnA3 reaction product, gamma-glutamylputrescine, was detected by HPLC/ESI-MS. In this work, the first step of polyamine utilization in S. coelicolor has been elucidated and the putative polyamine utilization pathway has been deduced based on the sequence similarity and transcriptional analysis of homologous genes expressed in the presence of polyamines.
Keywords: GS-like enzyme; GlnA3; Streptomyces coelicolor; gamma-glutamylpolyamine synthetase; nitrogen assimilation; polyamine utilization.
Figures




References
-
- Algranati I. D., Echandi G., Garcia-Patrone M., Gonzalez N. S., Goldemberg S. H. (1976). Polyamines, equilibrium between ribosomal particles and protein synthesis in bacteria. Arch. Biol. Med. Exp. 10 49–60. - PubMed
-
- Amin R., Franz-Wachtel M., Tiffert Y., Heberer M., Meky M., Ahmed Y., et al. (2016). Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145. Front. Mol. Biosci. 3:38 10.3389/fmolb.2016.00038 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

