Elucidating drug targets and mechanisms of action by genetic screens in mammalian cells
- PMID: 28487920
- PMCID: PMC5507204
- DOI: 10.1039/c7cc02349a
Elucidating drug targets and mechanisms of action by genetic screens in mammalian cells
Abstract
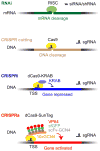
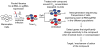
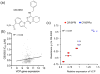
Phenotypic screening is a powerful approach to discover small molecules with desired effects on biological systems, which can then be developed into therapeutic drugs. The identification of the target and mechanism of action of compounds discovered in phenotypic screens remains a major challenge. This feature article describes the use of genetic tools to reveal drug targets and mechanisms in mammalian cells. Until recently, RNA interference was the method of choice for such studies. Here, we highlight very recent additions to the genetic toolkit in mammalian cells, including CRISPR, CRISPR interference, and CRISPR activation, and illustrate their usefulness for drug target identification.
Figures


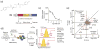
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

