A national study of the molecular epidemiology of HIV-1 in Australia 2005-2012
- PMID: 28489920
- PMCID: PMC5425008
- DOI: 10.1371/journal.pone.0170601
A national study of the molecular epidemiology of HIV-1 in Australia 2005-2012
Abstract
Introduction: Rates of new HIV-1 diagnoses are increasing in Australia, with evidence of an increasing proportion of non-B HIV-1 subtypes reflecting a growing impact of migration and travel. The present study aims to define HIV-1 subtype diversity patterns and investigate possible HIV-1 transmission networks within Australia.
Methods: The Australian Molecular Epidemiology Network (AMEN) HIV collaborating sites in Western Australia, South Australia, Victoria, Queensland and western Sydney (New South Wales), provided baseline HIV-1 partial pol sequence, age and gender information for 4,873 patients who had genotypes performed during 2005-2012. HIV-1 phylogenetic analyses utilised MEGA V6, with a stringent classification of transmission pairs or clusters (bootstrap ≥98%, genetic distance ≤1.5% from at least one other sequence in the cluster).
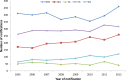
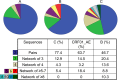
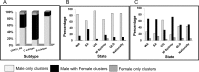
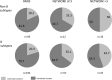
Results: HIV-1 subtype B represented 74.5% of the 4,873 sequences (WA 59%, SA 68.4%, w-Syd 73.8%, Vic 75.6%, Qld 82.1%), with similar proportion of transmission pairs and clusters found in the B and non-B cohorts (23% vs 24.5% of sequences, p = 0.3). Significantly more subtype B clusters were comprised of ≥3 sequences compared with non-B clusters (45.0% vs 24.0%, p = 0.021) and significantly more subtype B pairs and clusters were male-only (88% compared to 53% CRF01_AE and 17% subtype C clusters). Factors associated with being in a cluster of any size included; being sequenced in a more recent time period (p<0.001), being younger (p<0.001), being male (p = 0.023) and having a B subtype (p = 0.02). Being in a larger cluster (>3) was associated with being sequenced in a more recent time period (p = 0.05) and being male (p = 0.008).
Conclusion: This nationwide HIV-1 study of 4,873 patient sequences highlights the increased diversity of HIV-1 subtypes within the Australian epidemic, as well as differences in transmission networks associated with these HIV-1 subtypes. These findings provide epidemiological insights not readily available using standard surveillance methods and can inform the development of effective public health strategies in the current paradigm of HIV prevention in Australia.
Conflict of interest statement
Figures



References
-
- Roberts JD, Bebenek K, Kunkel T. The accuracy of reverse transcriptase from HIV-1. Science 1988; 242(4882): 1171–3. - PubMed
-
- Hemelaar J, Gouws E, Ghys PD, Osmanov S: WHO-UNAIDS Network for HIV Isolation and Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011; 25:679–89. doi: 10.1097/QAD.0b013e328342ff93 - DOI - PMC - PubMed
-
- Ivanov I, Beshkov D, Shankar A, Hanson D, Paraskevis D, Georgieva V et al. Detailed molecular epidemiologic characterization of HIV-1 infection in Bulgaria reveals broad diversity and evolving phylodynamics. PLoS One. 2013; 8(3):e59666 doi: 10.1371/journal.pone.0059666 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

