Integrated mRNA and microRNA transcriptome variations in the multi-tepal mutant provide insights into the floral patterning of the orchid Cymbidium goeringii
- PMID: 28490318
- PMCID: PMC5426072
- DOI: 10.1186/s12864-017-3756-9
Integrated mRNA and microRNA transcriptome variations in the multi-tepal mutant provide insights into the floral patterning of the orchid Cymbidium goeringii
Abstract
Background: Cymbidium goeringii is a very famous traditional orchid plant in China, which is well known for its spectacular and diverse flower morphology. In particular, the multi-tepal mutants have considerable ecological and cultural value. However, the current understanding of the molecular mechanisms of floral patterning and multi-tepal development is limited. In this study, we performed expression profiling of both microRNA (miRNA) and mRNA from wild-type and typical multi-tepal-mutant flowers of C. goeringii for the first time, to identify the genes and pathways regulating floral morphogenesis in C. goeringii.
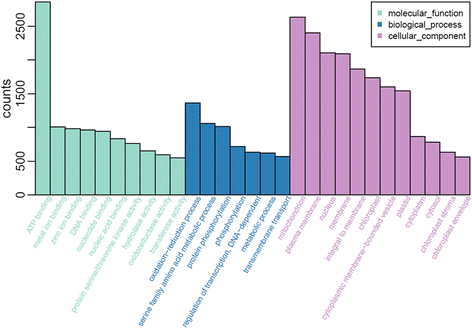
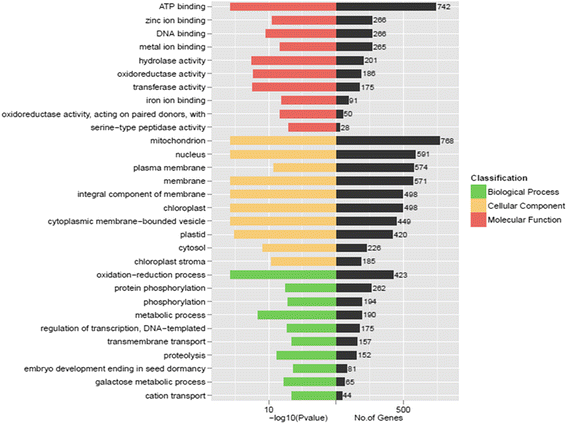
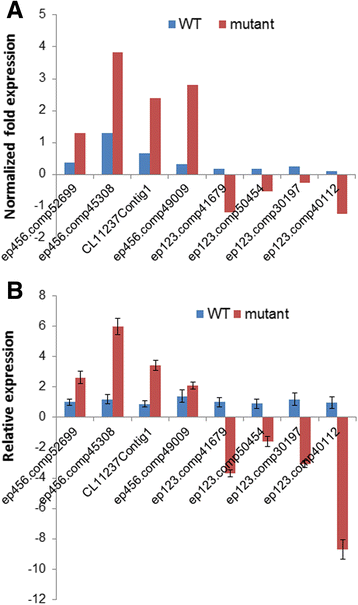
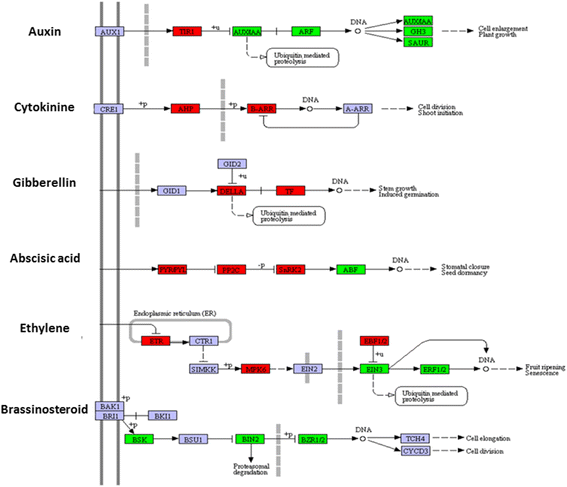
Results: Total clean reads of 98,988,774 and 100,188,534 bp were obtained from the wild-type and mutant library, respectively, and de novo assembled into 98,446 unigenes, with an average length of 989 bp. Among them, 18,489 were identified as differentially expressed genes between the two libraries according to comparative transcript profiling. The majority of the gene ontology terms and Kyoto Encyclopedia of Genes and Genomes pathway enrichment responses were for membrane-building and ploidy-related processes, consistent with the excessive floral organs and altered cell size observed in the mutant. There were 29 MADS-box genes, as well as a large number of floral-related regulators and hormone-responsive genes, considered as candidates regulating floral patterning of C. goeringii. Small RNA sequencing revealed 132 conserved miRNA families expressed in flowers of C. goeringii, and 11 miRNAs corresponding to 455 putative target genes were considered to be responsible for multi-tepal development. Importantly, integrated analysis of mRNA and miRNA sequencing data showed two transcription factor/microRNA-based genetic pathways contributing to the multi-tepal trait: well-known floral-related miR156/SPL and miR167/ARF regulatory modes involved in reproductive organ development; and the miR319/TCP4-miR396/GRF regulatory cascade probably regulating cell proliferation of the multi-tepal development.
Conclusions: Integrated mRNA and miRNA profiling data provided comprehensive gene expression information on the wild-type and multi-tepal mutant at the transcriptional level that could facilitate our understanding of the molecular mechanisms of floral patterning of C. goeringii. These data could also be used as an important resource for investigating the genetics of floral morphogenesis and various biological mechanisms of orchid plants.
Keywords: Cymbidium goeringii; Floral patterning; Floral transcriptome; MiR396; MicroRNA; Multi-tepal mutant.
Figures









References
-
- Du Puy DJ, Cribb P, Tibbs M. The Genus Cymbidium. Kew: Royal Botanic Gardens; 2007.
-
- Wu ZY, Raven PH, Hong DY. Flora of China. Beijing, St. Louis: Science Press and Missouri Botanical Garden Press; 2009.
-
- Chung MY, Chung MG. The breeding systems of Cremastra appendiculata and Cymbidium goeringii: high levels of annual fruit failure in two self-compatible orchids. Ann Bot Fenn. 2003;40(2):81–85.
-
- Rajkumari JD, Longjam RS. Orchid flower evolution. J Genet. 2005;84(1):81–4. - PubMed
-
- Yukawa T, Stern WL. Comparative vegetative anatomy and systematics of Cymbidium (Cymbidieae : Orchidaceae) Bot J Linn Soc. 2002;138(4):383–419. doi: 10.1046/j.1095-8339.2002.00038.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

