Genetic and Epidemiologic Trends of Norovirus Outbreaks in the United States from 2013 to 2016 Demonstrated Emergence of Novel GII.4 Recombinant Viruses
- PMID: 28490488
- PMCID: PMC5483924
- DOI: 10.1128/JCM.00455-17
Genetic and Epidemiologic Trends of Norovirus Outbreaks in the United States from 2013 to 2016 Demonstrated Emergence of Novel GII.4 Recombinant Viruses
Erratum in
-
Correction for Cannon et al., "Genetic and Epidemiologic Trends of Norovirus Outbreaks in the United States from 2013 to 2016 Demonstrated Emergence of Novel GII.4 Recombinant Viruses".J Clin Microbiol. 2019 Jun 25;57(7):e00695-19. doi: 10.1128/JCM.00695-19. Print 2019 Jul. J Clin Microbiol. 2019. PMID: 31239401 Free PMC article. No abstract available.
Abstract
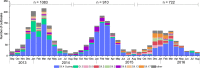
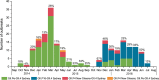
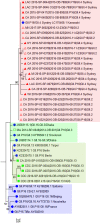
Noroviruses are the most frequent cause of epidemic acute gastroenteritis in the United States. Between September 2013 and August 2016, 2,715 genotyped norovirus outbreaks were submitted to CaliciNet. GII.4 Sydney viruses caused 58% of the outbreaks during these years. A GII.4 Sydney virus with a novel GII.P16 polymerase emerged in November 2015, causing 60% of all GII.4 outbreaks in the 2015-2016 season. Several genotypes detected were associated with more than one polymerase type, including GI.3, GII.2, GII.3, GII.4 Sydney, GII.13, and GII.17, four of which harbored GII.P16 polymerases. GII.P16 polymerase sequences associated with GII.2 and GII.4 Sydney viruses were nearly identical, suggesting common ancestry. Other common genotypes, each causing 5 to 17% of outbreaks in a season, included GI.3, GI.5, GII.2, GII.3, GII.6, GII.13, and GII.17 Kawasaki 308. Acquisition of alternative RNA polymerases by recombination is an important mechanism for norovirus evolution and a phenomenon that was shown to occur more frequently than previously recognized in the United States. Continued molecular surveillance of noroviruses, including typing of both polymerase and capsid genes, is important for monitoring emerging strains in our continued efforts to reduce the overall burden of norovirus disease.
Keywords: genetic recombination; genotypic identification; noroviruses.
Figures


References
-
- Wikswo ME, Kambhampati A, Shioda K, Walsh KA, Bowen A, Hall AJ. 2015. Outbreaks of acute gastroenteritis transmitted by person-to-person contact, environmental contamination, and unknown modes of transmission—United States, 2009–2013. MMWR Surveill Summ 64(SS12):1–16. doi: 10.15585/mmwr.ss6412a1. - DOI - PubMed
-
- Payne DC, Vinje J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, Wikswo M, Shirley SH, Hall AJ, Lopman B, Parashar UD. 2013. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med 368:1121–1130. doi: 10.1056/NEJMsa1206589. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

