The Cox1 C-terminal domain is a central regulator of cytochrome c oxidase biogenesis in yeast mitochondria
- PMID: 28490636
- PMCID: PMC5491776
- DOI: 10.1074/jbc.M116.773077
The Cox1 C-terminal domain is a central regulator of cytochrome c oxidase biogenesis in yeast mitochondria
Abstract
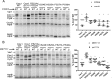
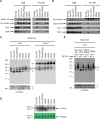
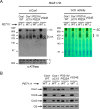
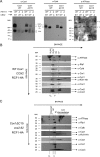
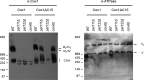
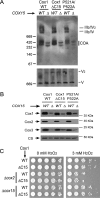
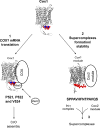
Cytochrome c oxidase (CcO) is the last electron acceptor in the respiratory chain. The CcO core is formed by mitochondrial DNA-encoded Cox1, Cox2, and Cox3 subunits. Cox1 synthesis is highly regulated; for example, if CcO assembly is blocked, Cox1 synthesis decreases. Mss51 activates translation of COX1 mRNA and interacts with Cox1 protein in high-molecular-weight complexes (COA complexes) to form the Cox1 intermediary assembly module. Thus, Mss51 coordinates both Cox1 synthesis and assembly. We previously reported that the last 15 residues of the Cox1 C terminus regulate Cox1 synthesis by modulating an interaction of Mss51 with Cox14, another component of the COA complexes. Here, using site-directed mutagenesis of the mitochondrial COX1 gene from Saccharomyces cerevisiae, we demonstrate that mutations P521A/P522A and V524E disrupt the regulatory role of the Cox1 C terminus. These mutations, as well as C terminus deletion (Cox1ΔC15), reduced binding of Mss51 and Cox14 to COA complexes. Mss51 was enriched in a translationally active form that maintains full Cox1 synthesis even if CcO assembly is blocked in these mutants. Moreover, Cox1ΔC15, but not Cox1-P521A/P522A and Cox1-V524E, promoted formation of aberrant supercomplexes in CcO assembly mutants lacking Cox2 or Cox4 subunits. The aberrant supercomplex formation depended on the presence of cytochrome b and Cox3, supporting the idea that supercomplex assembly factors associate with Cox3 and demonstrating that supercomplexes can be formed even if CcO is inactive and not fully assembled. Our results indicate that the Cox1 C-terminal end is a key regulator of CcO biogenesis and that it is important for supercomplex formation/stability.
Keywords: Cox1; Mss51; cytochrome c oxidase (complex IV); mitochondria; mitochondrial DNA (mtDNA); supercomplex; translation; yeast.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae.J Biol Chem. 2011 Jan 7;286(1):555-66. doi: 10.1074/jbc.M110.188805. Epub 2010 Nov 10. J Biol Chem. 2011. PMID: 21068384 Free PMC article.
-
The carboxyl-terminal end of Cox1 is required for feedback assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria.J Biol Chem. 2010 Nov 5;285(45):34382-9. doi: 10.1074/jbc.M110.161976. Epub 2010 Aug 31. J Biol Chem. 2010. PMID: 20807763 Free PMC article.
-
A Novel Function of Pet54 in Regulation of Cox1 Synthesis in Saccharomyces cerevisiae Mitochondria.J Biol Chem. 2016 Apr 22;291(17):9343-55. doi: 10.1074/jbc.M116.721985. Epub 2016 Feb 29. J Biol Chem. 2016. PMID: 26929411 Free PMC article.
-
Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core.Biochim Biophys Acta. 2012 Jun;1817(6):883-97. doi: 10.1016/j.bbabio.2011.09.005. Epub 2011 Sep 16. Biochim Biophys Acta. 2012. PMID: 21958598 Free PMC article. Review.
-
Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane.Gene. 2005 Jul 18;354:43-52. doi: 10.1016/j.gene.2005.03.017. Gene. 2005. PMID: 15905047 Review.
Cited by
-
Mitochondrial proteins and congenital birth defect risk: a mendelian randomization study.BMC Pregnancy Childbirth. 2025 Apr 14;25(1):444. doi: 10.1186/s12884-025-07562-8. BMC Pregnancy Childbirth. 2025. PMID: 40229705 Free PMC article.
-
Mss51 deletion enhances muscle metabolism and glucose homeostasis in mice.JCI Insight. 2019 Oct 17;4(20):e122247. doi: 10.1172/jci.insight.122247. JCI Insight. 2019. PMID: 31527314 Free PMC article.
-
The Complexity of Mitochondrial Complex IV: An Update of Cytochrome c Oxidase Biogenesis in Plants.Int J Mol Sci. 2018 Feb 27;19(3):662. doi: 10.3390/ijms19030662. Int J Mol Sci. 2018. PMID: 29495437 Free PMC article. Review.
-
Interplay between Mitochondrial Protein Import and Respiratory Complexes Assembly in Neuronal Health and Degeneration.Life (Basel). 2021 May 11;11(5):432. doi: 10.3390/life11050432. Life (Basel). 2021. PMID: 34064758 Free PMC article. Review.
-
Arabidopsis mtHSC70-1 physically interacts with the Cox2 subunit of cytochrome c oxidase.Plant Signal Behav. 2020;15(2):1714189. doi: 10.1080/15592324.2020.1714189. Epub 2020 Jan 14. Plant Signal Behav. 2020. PMID: 31933409 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

