Transcribing the connectome: roles for transcription factors and chromatin regulators in activity-dependent synapse development
- PMID: 28490640
- PMCID: PMC5539463
- DOI: 10.1152/jn.00067.2017
Transcribing the connectome: roles for transcription factors and chromatin regulators in activity-dependent synapse development
Abstract
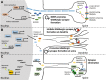
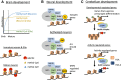
The wiring of synaptic connections in the developing mammalian brain is shaped by both intrinsic and extrinsic signals. One point where these regulatory pathways converge is via the sensory experience-dependent regulation of new gene transcription. Recent studies have elucidated a number of molecular mechanisms that allow nuclear transcription factors and chromatin regulatory proteins to encode aspects of specificity in experience-dependent synapse development. Here we review the evidence for the transcriptional mechanisms that sculpt activity-dependent aspects of synaptic connectivity during postnatal development and discuss how disruption of these processes is associated with aberrant brain development in autism and intellectual disability.
Keywords: activity-dependent synaptic plasticity; autism; chromatin; synapse development; transcription.
Copyright © 2017 the American Physiological Society.
Figures

References
-
- Ataman B, Boulting GL, Harmin DA, Yang MG, Baker-Salisbury M, Yap EL, Malik AN, Mei K, Rubin AA, Spiegel I, Durresi E, Sharma N, Hu LS, Pletikos M, Griffith EC, Partlow JN, Stevens CR, Adli M, Chahrour M, Sestan N, Walsh CA, Berezovskii VK, Livingstone MS, Greenberg ME. Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 539: 242–247, 2016. doi: 10.1038/nature20111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

